SilcsBio Suites and Features
Overview
SilcsBio offers a range of tools for computer-aided drug design. Our users include academic and government research institues as well as large pharmaceutical companies. Our current offerings are:
Small Molecule (SM) Suite
The SM Suite provides tools for end-to-end small molecule ligand drug design. Map chemical functional group affinity patterns of your target protein, identify ligand binding sites, virtually screen our databases of up to 1.9 billion compounds with target-based pharmacophore models, dock your ligands of interest, and optimize your lead compound to design your small molecule drug.
Covalent Suite
The Covalent Suite streamlines your covalent drug design process by combining physics-based chemical functional group affinity patterns and machine learning models. Identify the most reactive residues within your protein, predict the optimal warheads for your target residue, model the binding of your covalent inhibitor, and optimize your covalent inhibitor to maximize its potency.
SM+ Suite
Combine the power of the SM and Covalent Suites with the SM+ Suite. The SM+ Suite provides users with all the features in the SM and Covalent Suites in one package.
Biologics Suite
The Biologics Suite provides tools that rationalize the selection of excipients for formulating your protein-based therapeutic. Predict regions of your therapeutic protein prone to self-aggregation, map chemical functional group affinity patterns for your protein, predict excipient interaction sites, and characterize excipient-protein interactions in relation to protein-protein aggregation to identify excipients that stabilize the active fold of your protein and prevent aggregation thereby reducing viscosity for your biologic’s optimal formulation.
SSFEP Suite
The SSFEP Suite leverages the single step free energy pertubation (SSFEP) method to circumvent the large computational cost of free energy perturbation (FEP) while maintaining comparible accuracy for small functional group modifications. Introduce functional group modifications to your parent ligand and rapidly evaluate hundreds of potential modifications involving multiple sites on your parent ligand for your target protein.
CGenFF Suite
The CGenFF Suite enables accurate molecular dynamics (MD) simulations of biomolecular systems. Rapidly generate force field (FF) topologies and parameters for novel ligands on the fly, including covalently bonded ligands, run standard molecular dynamics simulations, and perform structural and energetic analyses or your biomolecular system.
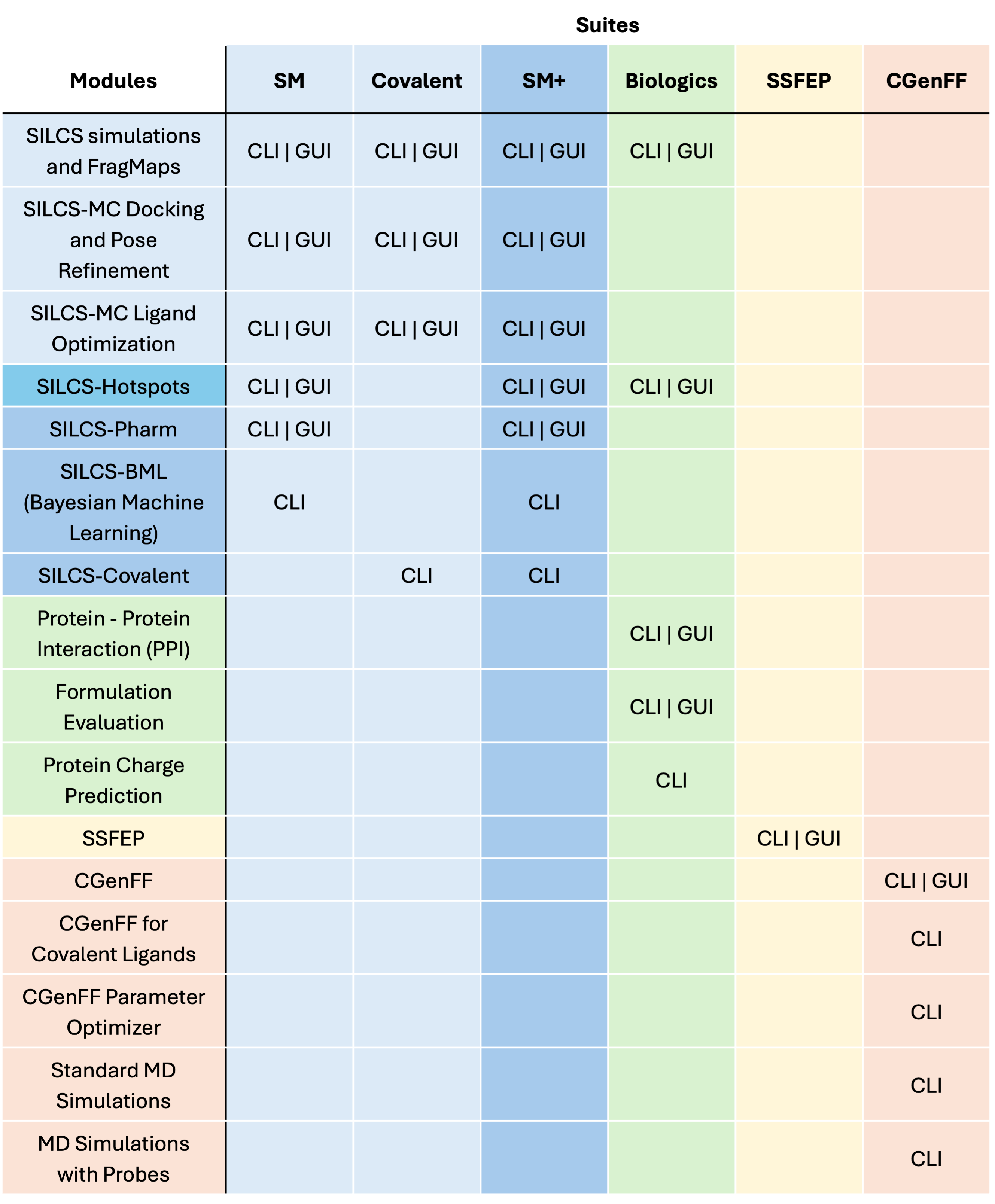
For more details on what features are avalable for each suite, please see Suite Feature and Module Comparison.
Suite Feature and Module Comparison
The suites and their included features and modules are listed in the table below. Features marked with “CLI” are available only in the command line interface, and features marked with “CLI | GUI” are available in both the command line interface (CLI) and the SilcsBio graphical user interface (GUI). If you have any questions about the suites or any particular feature, please contact info@silcsbio.com.