Analysis of Ligand Substructures
The SilcsBio software provides a utility through the command line interface (CLI) to calculate the properties of substructures within SILCS-MC docked ligands.
python $SILCSBIODIR/utils/python/delta_lgfe_analysis.py --minconfpdb <minconfpdb directory>
Required parameters:
Path to directory containing SILCS-MC docked ligands:
--minconfpdb <minconfpdb directory>
Path to parent structure SD file:
--parent <parent molecule SD file>
Optional parameters:
Path to output CSV file:
--output <output csv file>
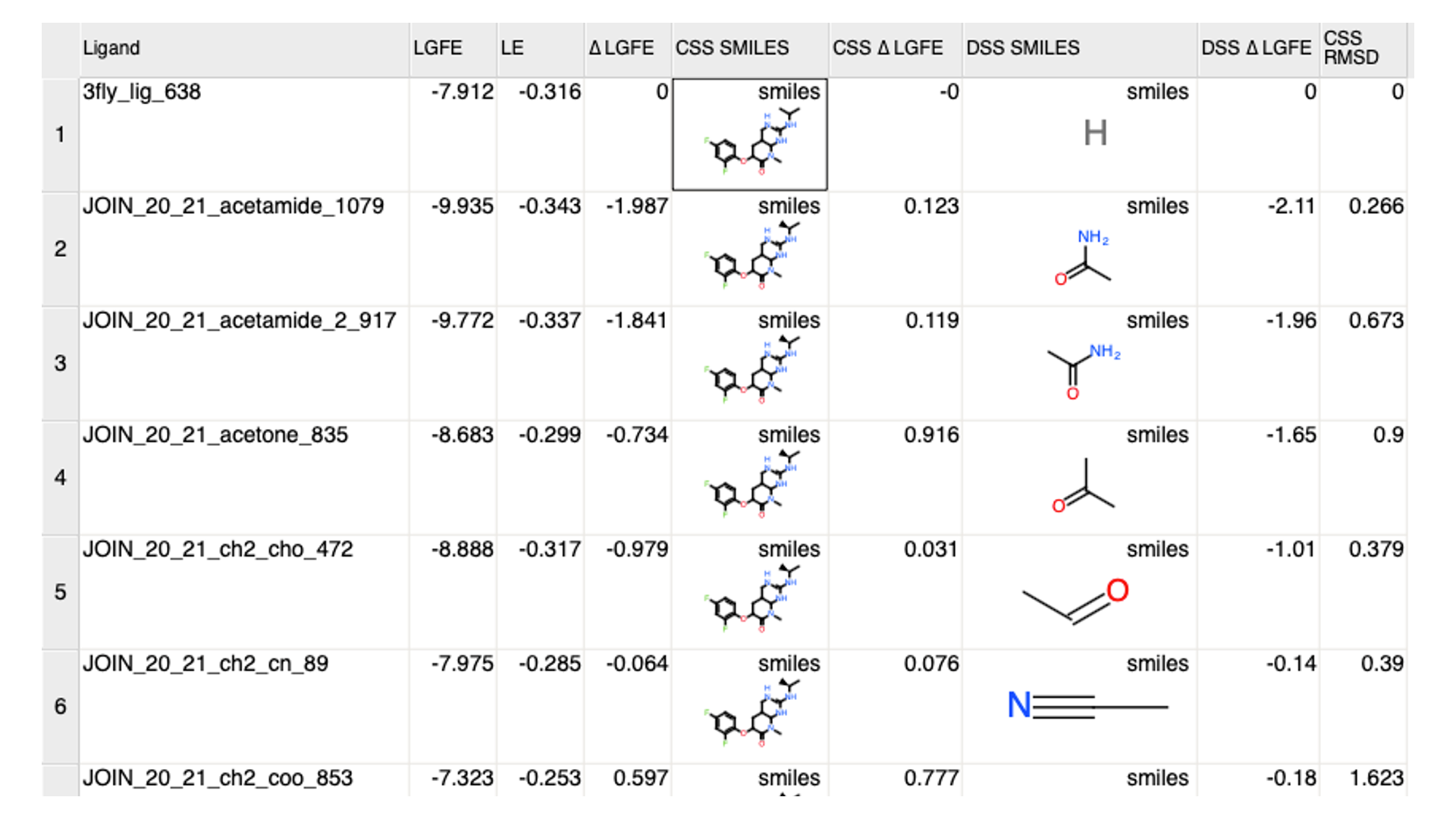
After running the above command, a csv (delta_lgfe_analysis.csv by default) will be
created. The csv file will list the
ligand name (
Ligand, first column),total LGFE (
LGFE, second column),LE (
LE, third column),change in total LGFE from the parent ligand (
dLGFE, fourth column),SMILES of the common substructure (
CSS SMILES, fifth column),change in LGFE contribution of the common substructure (
CSS dLGFE, sixth column),SMILES of the modification or different substructure (
DSS SMILES, seventh column),change in LGFE contribution by the modification or different substructure (
DSS dLGFE, eight column), andRMSD of the common substructure in reference to the parent ligand (
CSS RMSD, nineth column).
The output csv file is compatible with cheminformatics softwares such as MOE or Molsoft for viewing and further processing. Below is the output visualized in Molsoft with SMILES converted to 2D structures: