SILCS-MC: Docking and Pose Refinement¶
SILCS-MC Background¶
The power of SILCS lies in the ability to use FragMaps to rapidly evaluate binding of diverse ligands to a target. SILCS-MC is Monte-Carlo (MC) sampling of ligands in translational, rotational, and torsional space in the field of FragMaps. MC sampling uses CGenFF intramolecular energies and the Ligand Grid Free Energy (LGFE), which is the sum of atomic Grid Free Energies (GFEs). The Exclusion Map prevents ligand sampling where no probe or water molecules visited during SILCS simulations. SILCS-MC allows for rapid conformational sampling of the ligand while accounting for protein flexibility in a mean-field-like fashion since ligand affinity and volume exclusion are embedded in the combination of FragMaps and the Exclusion Map. Final ligand scoring is based on the LGFE score [9][22].
SILCS-MC can be used to generate and score binding poses for a ligand to a target using SILCS FragMaps that have been previously computed for that target (see SILCS: Site Identification by Ligand Competitive Saturation). This can readily be done for a single ligand or a database of ligands. Two default conformational sampling protocols are available, “docking” and “pose refinement,” as described in detail below. The docking protocol is useful when no information is available about the binding pose, as it entails extensive translational, rotational, and intramolecular conformational sampling. The pose refinement protocol is useful when a reasonable starting pose for the ligand is available, for example to re-score poses output by another high-throughput in silico screening tool. Instructions for developing custom SILCS-MC ligand conformational sampling protocols are provided at the end of this chapter.
Running SILCS-MC docking¶
Docking is exhaustive sampling of a ligand’s conformation in a given pocket to determine its most favorable orientation and internal geometry as defined by LGFE scoring. The pocket is predefined as a 10 Å sphere and the center of the pocket is taken as either the center of the supplied ligand molecule coordinates or explicitly given by the user. This protocol entails five independent MC runs with the ligand.
This protocol is recommended for ligands with diverse chemotypes and unknown binding poses. When the pose of a parent ligand is known and SILCS-MC evaluations are to be performed over a congeneric series, the pose refinement protocol is recommended instead (see below).
Running SILCS-MC docking from the SilcsBio GUI¶
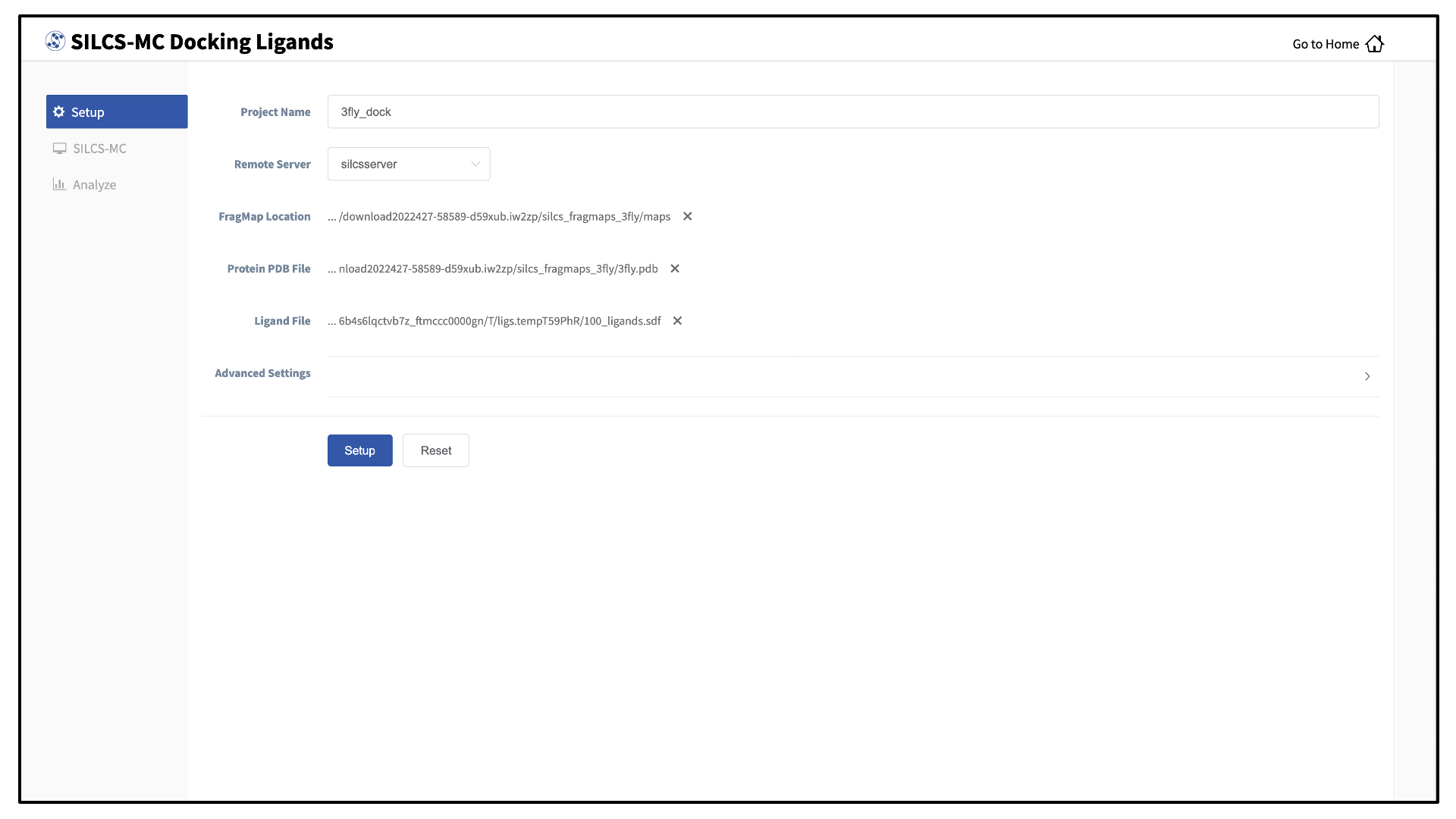
Select New SILCS-MC Docking project from the Home page.
Enter a project name and select the remote server where the SILCS-MC docking jobs will run. Also, provide FragMap and protein input files. You may choose these files from the computer where you are running the SilcsBio GUI (“localhost”) or from any server you have previously configured, as described in File and directory selection. You will additionally need to provide a “Ligand SDF File” that contains the database of ligands to be docked.
Warning
Ligands in the “Ligand SDF File” must include all hydrogens, including pH-appropriate (de)protonations, and must have reasonable three dimensional conformations.
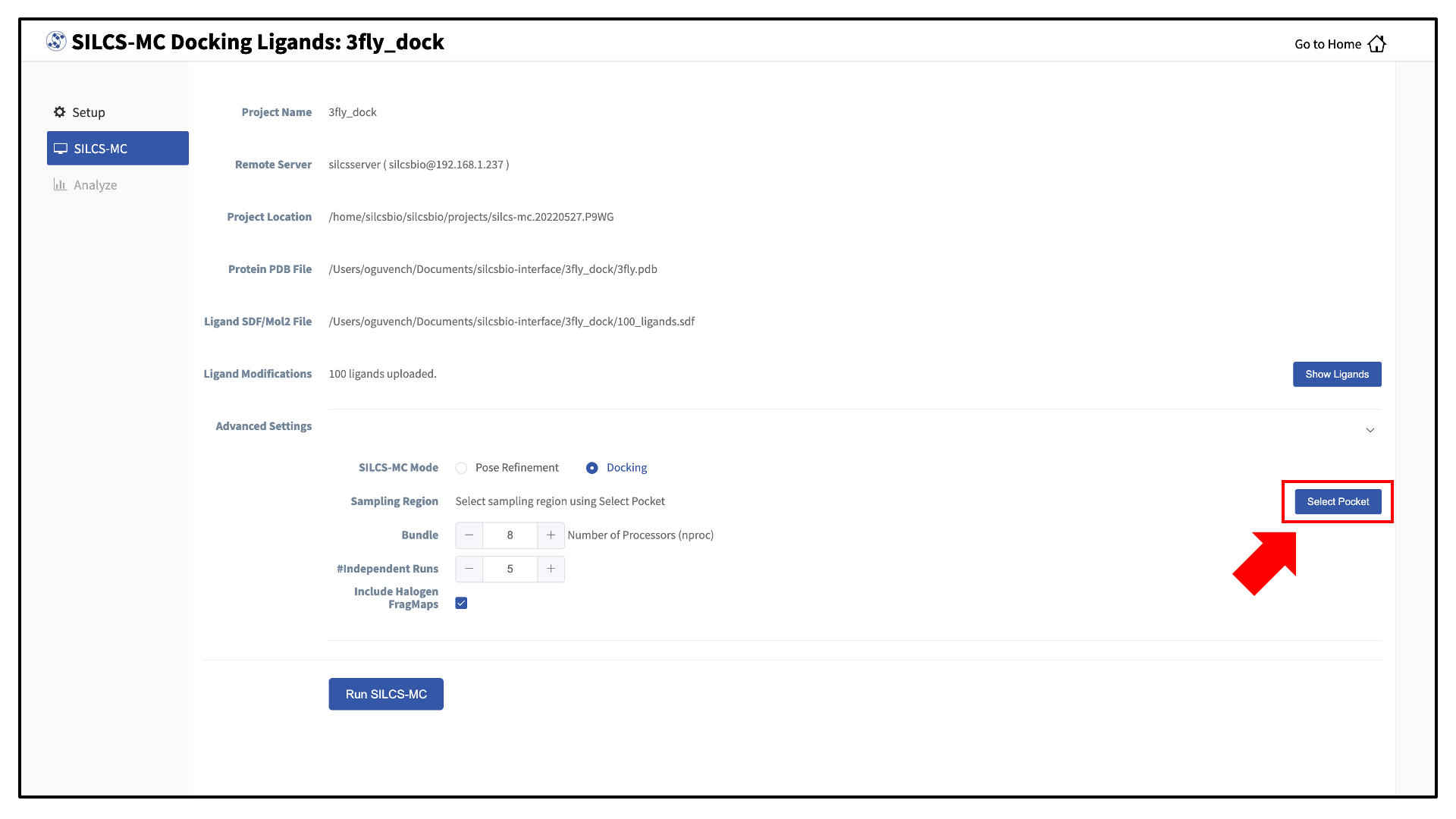
Once all information is entered correctly, press the “Setup” button at the bottom of the page. The page will update to list the number of ligands and show options for the sampling protocol (“Docking” or “Pose Refinement”) and the sampling region. Select the “Docking” option and then press the “Select Pocket” button.
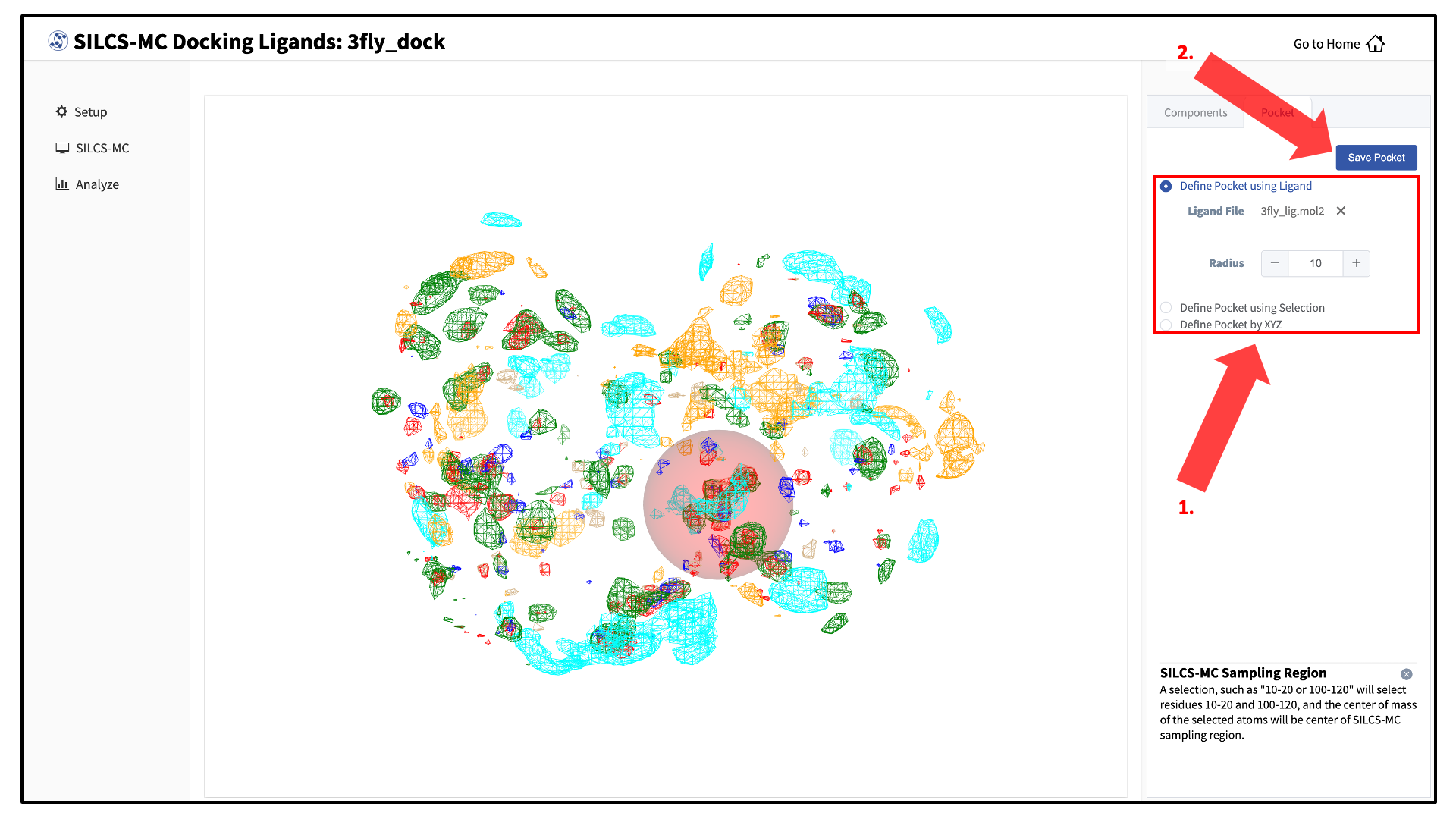
The GUI will now be showing the protein molecular graphic in the center pane. On the right-hand side in the “Pocket” tab, you can define the pocket center based on the center-of-geometry of a ligand pose (“Define Pocket using Ligand”), or a target residue selection (“Define Pocket using Selection”), or by directly entering an x, y, z coordinate (“Define Pocket by XYZ”). You will also need to choose a radius (default value “10”) to complete the definition of the spherical pocket. If it is difficult to see the spherical pocket definition in the center pane, hide the protein surface representation. Click on the “Save Pocket” button and the “OK” acknowledgement to continue.
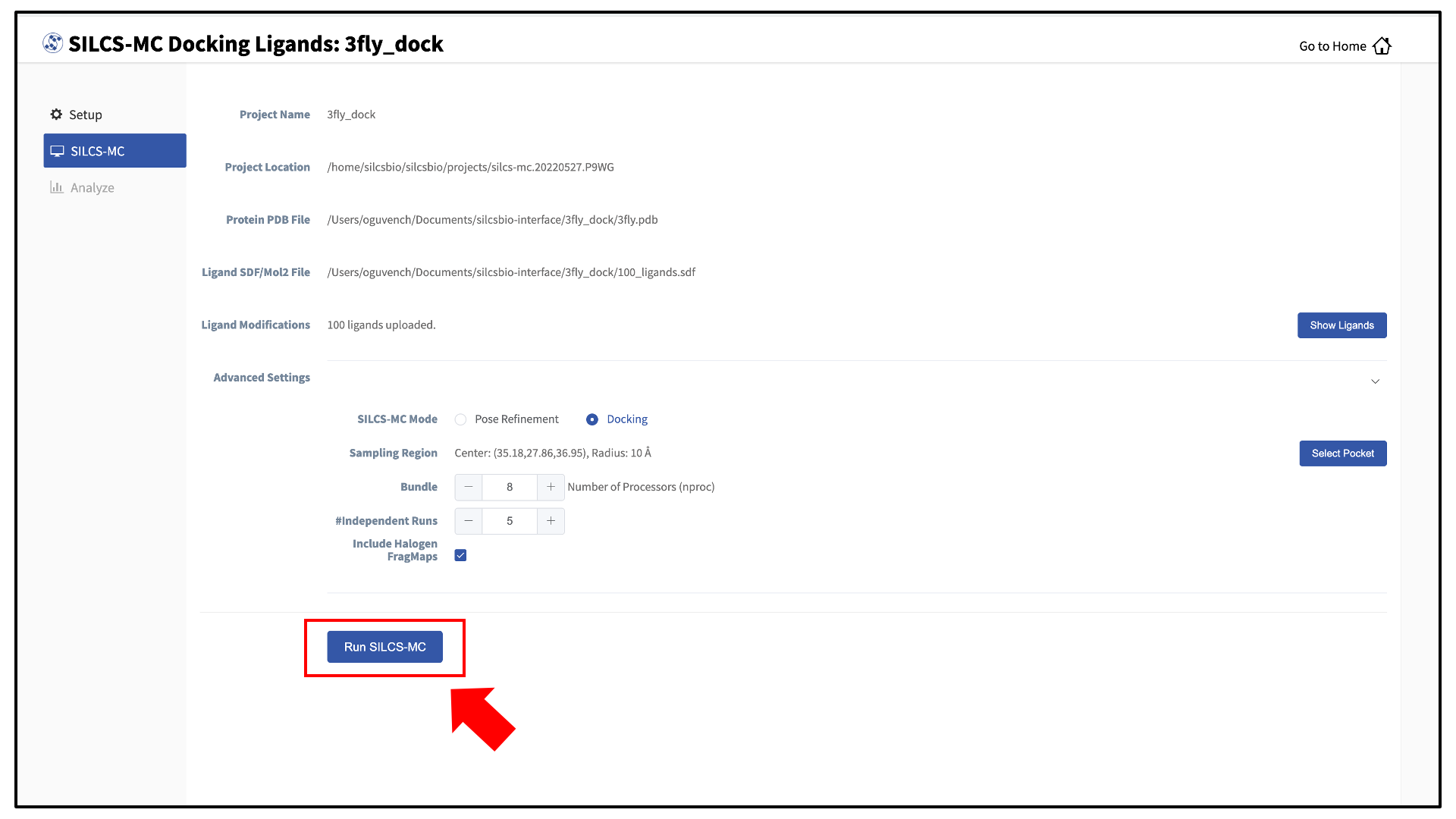
You will be returned to the previous screen, which now includes “Sampling Region” information consisting of the spherical pocket center and radius.
Tip
If you ran Halogen SILCS simulations for your target, you can include the Halogen SILCS FragMaps in the SILCS-MC posing and scoring by checking the “Include Halogen FragMaps” box.
You may now click the “Run SILCS-MC” button to start the SILCS-MC docking. Doing so will submit jobs to the remote server job schedular and list them in a pop-over window.
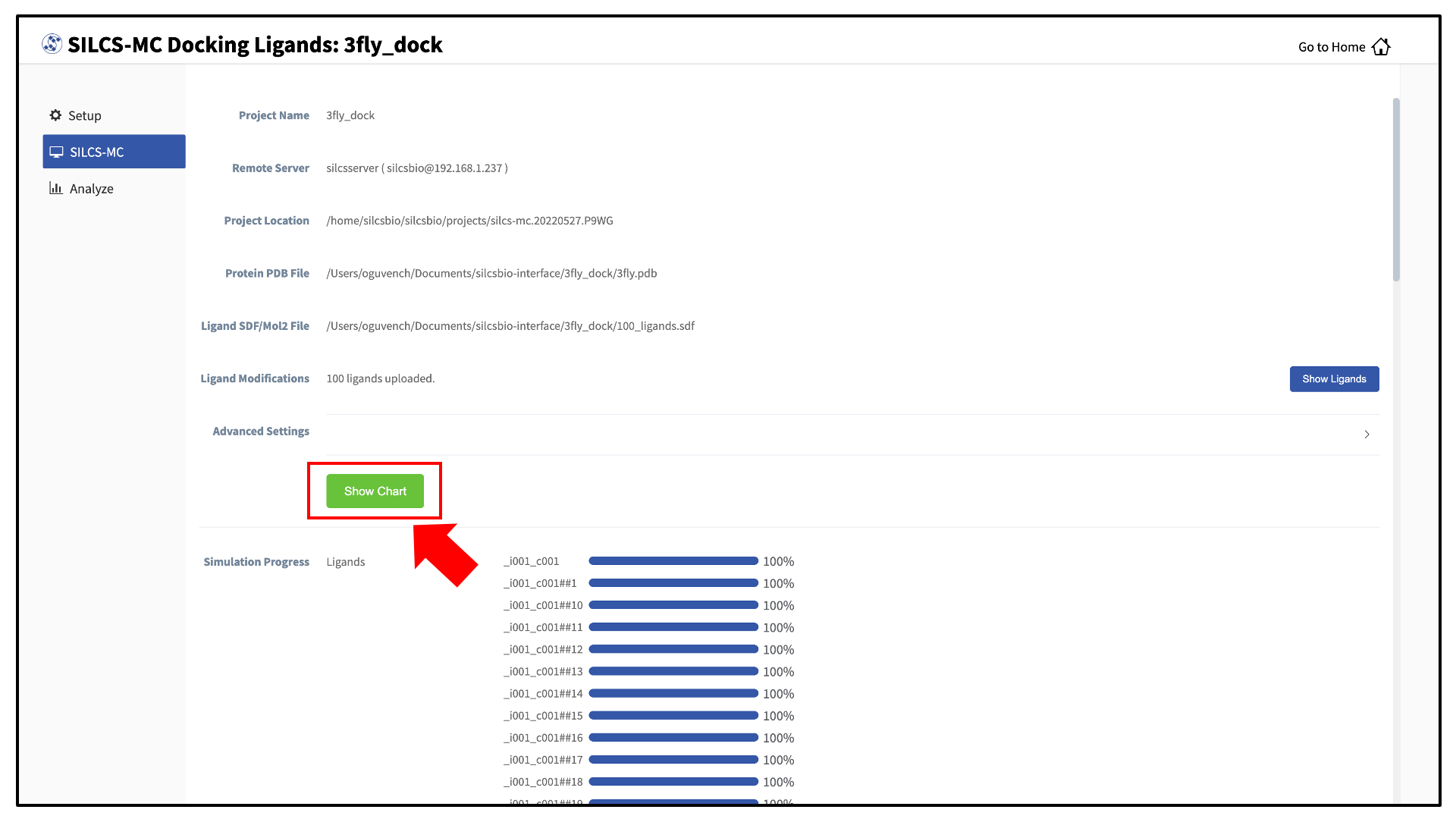
Once all jobs are submitted, you may click on the “Setup Successful” button to dismiss the pop-over window and return to the previous screen, which will now show a “Simulation Progress” section. You may update this section by scrolling to the bottom of the screen and clicking the “Refresh” button. This will update the progress bars for all of the ligands being docked.
Once progress bars for all ligands reach 100%, click on the “Show Chart” button above the “Simulation Progress” section to proceed.
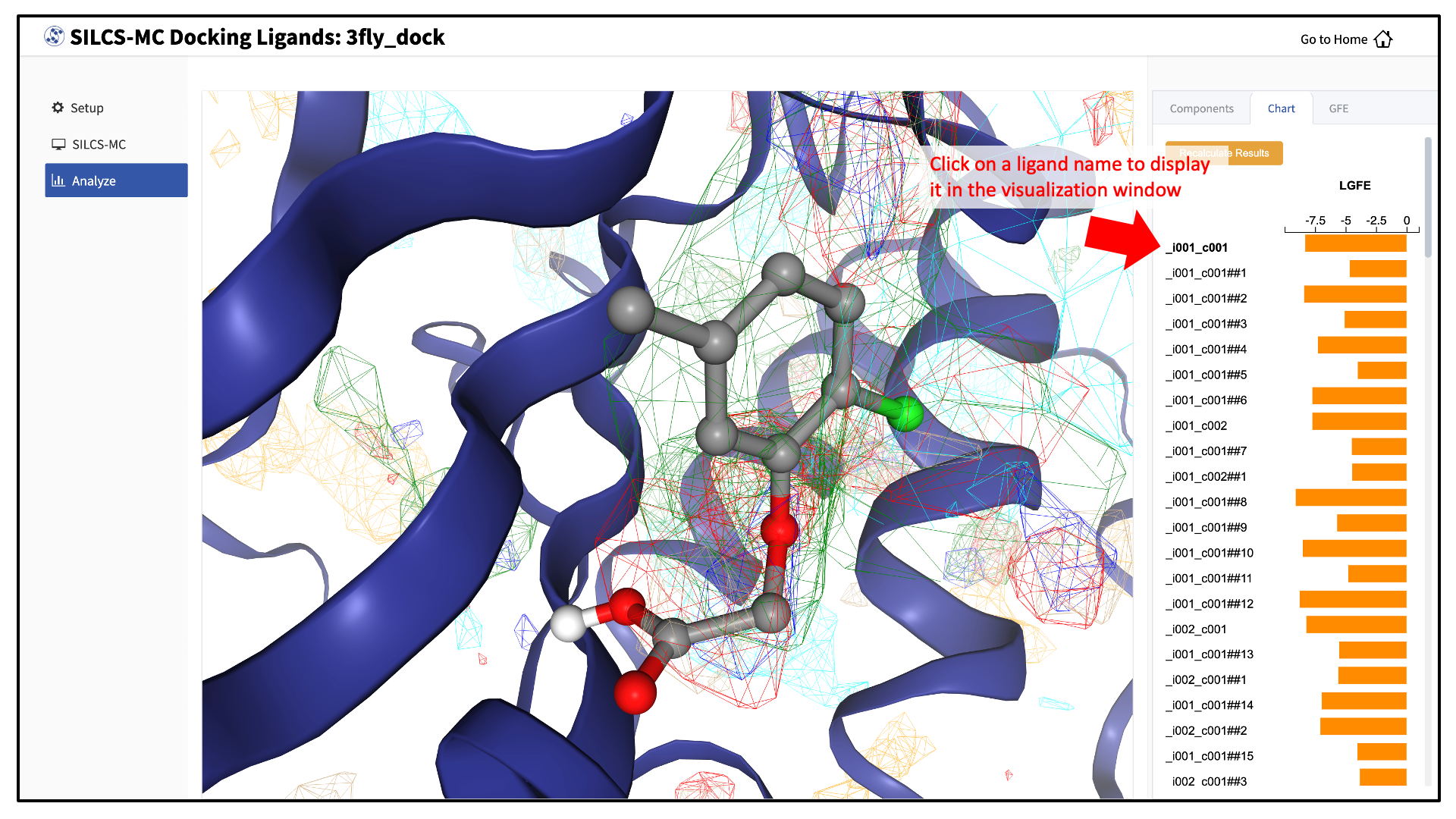
Upon successful completion of this command in the pop-over window, you may click on the green “Data collection finished” button to return to the GUI. A new tab, labeled “Chart” will have been created in the right-hand panel. Under that tab, clicking on a ligand name will display that ligand.
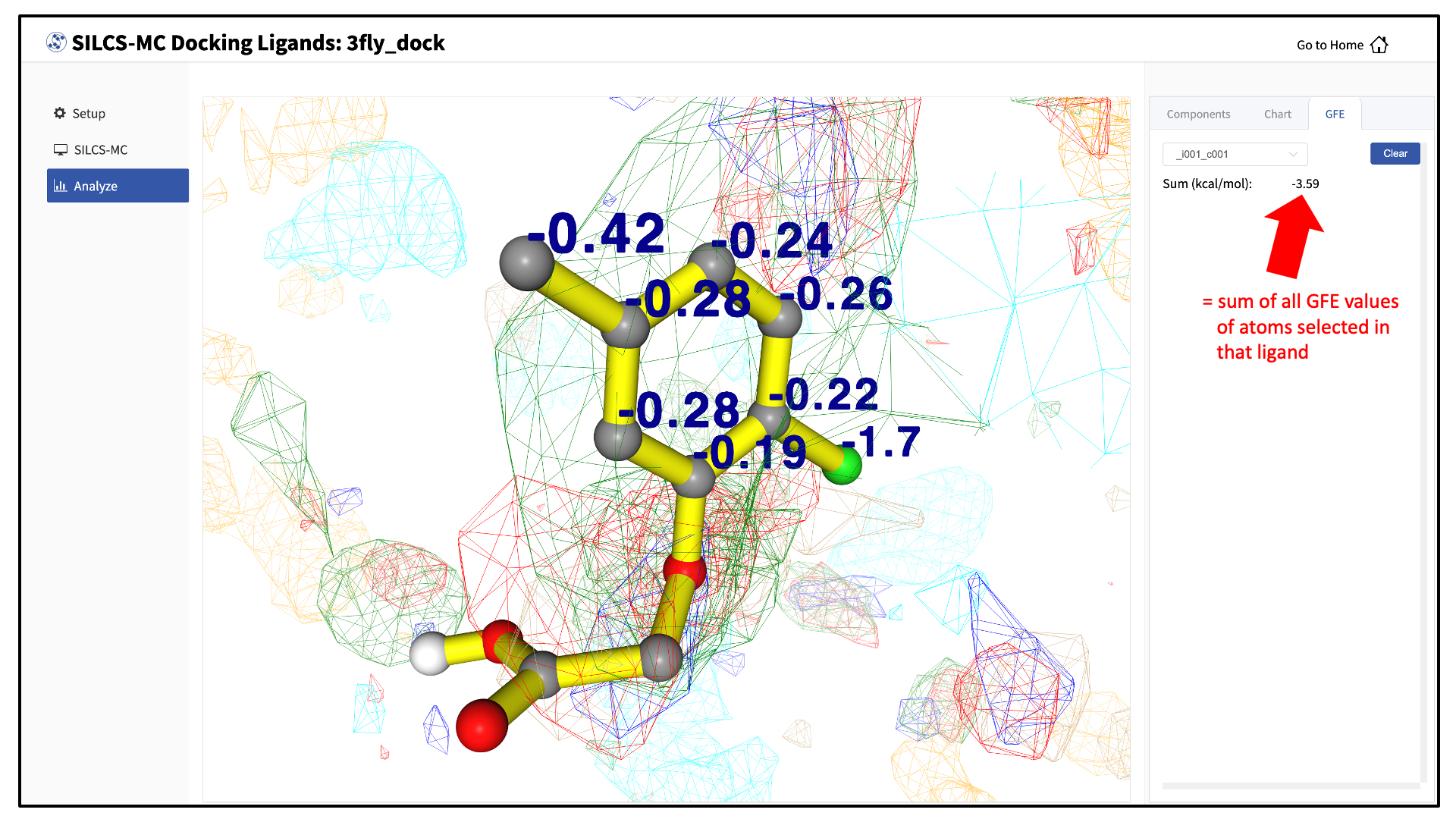
Choosing the “GFE” tab in the right-hand panel will allow you to click on individual atoms within a ligand to see their atomic GFE values. The “GFE” tab will also display the sum of all GFE values of atoms selected in that ligand.
Running SILCS-MC docking from the command line interface¶
To set up and run SILCS-MC docking from the command line interface, create a directory containing all the ligands to be evaluated. Each ligand can be stored as a separate SDF or Mol2 file. Alternatively, all the ligands can be combined into a single SDF file.
${SILCSBIODIR}/silcs-mc/1_run_silcsmc_exhaustive \
prot=<prot pdb> \
ligdir=<directory containing ligand mol2/sdf> \
mapsdir=<directory containing SILCS FragMaps> \
center="x,y,z"
Note
.sdf, .sd, or .mol2 files can be placed in the ligdir
directory, and SILCS-MC will read a single molecule from each file.
Note that if a file contains multiple molecules, use of the ligdir
option will result in only the first molecule in the file being processed.
If you have an SDF file with multiple molecules in it, replace
ligdir=<directory containing ligand mol2/sdf> with
sdfile=<path to sdfile> to process all molecules in the file.
Warning
Ligands, regardless of file format, must include all hydrogens, including pH-appropriate (de)protonations, and must have reasonable three dimensional conformations.
If halogen FragMaps have been generated (see SILCS simulation setup with halogen probes),
they can be included in the SILCS-MC calculation to improve the scoring of
halogen-containing compounds. To do so, add the halogen=true option to
the 1_run_silcsmc_exhaustive command.
SILCS-MC docking protocol details¶
Docking spawns five independent single-core serial jobs per ligand.
Each run involves a maximum of 250 cycles and a minimum of 50 cycles of
Monte Carlo/Simulated Annealing (MC/SA) sampling of the
ligand within the defined 10 Å sphere. Each of these 250 cycles consists of
10,000 steps of MC at a high temperature followed by 40,000 steps of
SA towards a lower temperature. At the beginning of
each cycle, the ligand will be reoriented within the predefined sphere. When
no sphere center is defined by the user, the ligand orientation in the input
SDF or Mol2 file will be used as the starting pose for each cycle.
The MC sampling has three types of moves: i) molecular translations with a
maximum step size of 1 Å, ii) molecular rotation with a maximum step size of
180 degrees, and iii) intramolecular dihedral rotations with a maximum
step size of 180 degrees. For intramolecular dihedral rotations, only the
rotatable dihedral angles are selected for MC moves. The lowest LGFE scoring
pose from the MC sampling is used as starting pose in the following SA
sampling. The SA sampling also involves the same three types of moves, but
with a smaller step size compared to the MC sampling: i) molecular
translations with a maximum step size of 0.2 Å, ii) molecular rotation with a
maximum step size of 9 degrees and, iii) intramolecular dihedral rotations with
a maximum step size of 9 degrees. The lowest LGFE scoring pose from the SA is
saved in a multi-frame PDB file: 3_silcsmc/<run>/pdb/<lig>_mc.<run>.pdb.
In each run, after a minimum of 50 MC/SA cycles, if the LGFE score difference between the top three poses (defined by lowest LGFE scores) are less than 0.5 kcal/mol, then that run is considered converged and terminated. If the top three scored poses are separated by more 0.5 than kcal/mol, the MC/SA procedure continues either until the convergence criterion is met or until a maximum of 250 MC/SA cycles have been completed.
Running SILCS-MC pose refinement¶
The pose refinement protocol is designed to limit conformational sampling near the ligand input pose supplied by the user. Pose refinement is appropriate when there is high confidence in the input parent ligand pose and SILCS-MC evaluations are to be performed over a congeneric series. The sphere center for the pocket definition is the center-of-geometry from the input ligand pose.
Running SILCS-MC pose refinement from the SilcsBio GUI¶
Please see the previous section on running SILCS-MC docking from the SilcsBio
GUI, and, in Step 3, select the “Pose Refinement” option. Note that there
will be no “Select Pocket” step, as pose refinement assigns the pocket center
based on the center-of-geometry of the input ligand. For multiple input
ligands in a single .sdf file, each ligand will have its own
center-of-geometry used to define the pocket center.
Running SILCS-MC pose refinement from the command line interface¶
To set up and run SILCS-MC pose refinement, create a directory containing all
the ligands to be evaluated. Each ligand can be stored as a separate .mol2
or .sdf file. Alternatively, all the ligands can be combined into a single
.sdf file.
${SILCSBIODIR}/silcs-mc/1_run_silcsmc_local prot=<prot pdb> \
ligdir=<directory containing ligand mol2/sdf> \
mapsdir=<directory containing SILCS FragMaps>
Note
.sdf, .sd, or .mol2 files can be placed in the ligdir
directory, and SILCS-MC will read a single molecule from each file.
Note that if a file contains multiple molecules, use of the ligdir
option will result in only the first molecule in the file being processed.
If you have an SDF file with multiple molecules in it, replace
ligdir=<directory containing ligand mol2/sdf> with
sdfile=<path to sdfile> to process all molecules in the file.
Warning
Ligands, regardless of file format, must include all hydrogens, including pH-appropriate (de)protonations, and must have reasonable three dimensional conformations.
If halogen FragMaps have been generated (see SILCS simulation setup with halogen probes),
they can be included in the SILCS-MC calculation to improve the scoring of
halogen-containing compounds. To do so, add the halogen=true option to
the 1_run_silcsmc_local command.
SILCS-MC pose refinement protocol details¶
Pose refinement spawns five independent single-core serial jobs per
ligand, and each run involves a maximum of 10 cycles of MC/SA sampling
of the ligand within a 1 Å sphere. The center of the sphere is defined
as the center-of-geometry of the input ligand pose. Each of cycles
consists of 100 steps of MC at high temperature followed by 1000 steps
of SA towards a lower temperature. At the beginning of each cycle, the
ligand orientation/conformation will be reset to the one found in the
input file. MC sampling moves are: i) molecular translation with a
maximum step size of 1 Å, ii) molecular rotation with a maximum step
size of 180 degrees, and iii) intramolecular dihedral rotation with a
maximum step size of 180 degrees. For intramolecular dihedral rotation,
only the rotatable dihedral angles are selected for MC moves. The lowest
LGFE scoring pose from the MC sampling is used as starting pose in the
following SA sampling. SA sampling moves are smaller than for the MC
phase: i) molecular translation with a maximum step size of 0.2 Å, ii)
molecular rotation with a maximum step size of 9 degrees, iii)
intramolecular dihedral rotation with a maximum step size of 9 degrees.
The lowest LGFE scoring pose from the SA is saved in the multi-frame PDB
file 3_silcsmc/<run>/pdb/<lig>_mc.<run>.pdb.
Best-pose retrieval¶
Once the SILCS-MC simulation is finished, retrieve the LGFE scores for each ligand subjected to SILCS-MC using:
${SILCSBIODIR}/silcs-mc/2_calc_lgfe_min_avg_sd ligdir=<directory containing lig mol2>
Note
If you used sdfile=<path to sdfile> option in the previous step, use
sdfile=<path to sdfile> in this step as well, instead of ligdir
option.
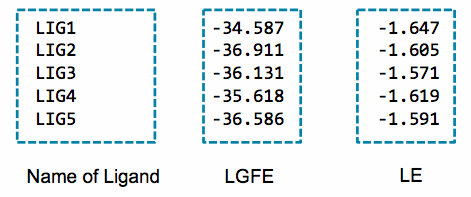
The above command will collect the SILCS-MC result and create
3_silcsmc/lgfe.csv file, which contains the best LGFE scores from
SILCS-MC sampling per ligand molecule. The command will collect top 10
best scoring poses and put them in 3_silcsmc/minconfpdb/ folder in
SDF format.
Note
To change the number of poses, add npose=<number> at the end of
2_calc_lgfe_min_avg_sd command.
To output PDB file format for best scoring poses, add pdb=true at
the end of 2_calc_lgfe_min_avg_sd command.
An example of the output of this script is:
An alternative to the LGFE score is the ligand efficiency (LE). The LE is calculated as the LGFE score divided by the number of heavy atoms in each ligand.
User-defined protocols¶
In addition to the default docking and pose refinement protocols, users
can define their own SILCS-MC protocols. To do so, copy
${SILCSBIODIR}/templates/silcs-mc/params_custom.tmpl to the location where
you intend to run your custom SILCS-MC protocol. Edit this copy to reflect your
customization; see below for a detailed description of user-definable
parameters. Parameter values in angle brackets in this file, such as
<SILCSBIODIR>, will be replaced automatically at runtime where possible.
After you have edited params_custom.tmpl, use the following command to set
up and run SILCS-MC. Each input ligand can be stored as a separate SDF or Mol2
file. Alternatively, all input ligands can be combined in a single SDF file.
${SILCSBIODIR}/silcs-mc/1_run_silcsmc_custom prot=<prot pdb> \
ligdir=<directory containing ligand sdf/mol2> \
mapsdir=<directory containing SILCS FragMaps> \
paramsfile=<params_custom.tmpl>
The number of runs that will be spawned can also modified with the command-line
parameter totruns:
${SILCSBIODIR}/silcs-mc/1_run_silcsmc_custom prot=<prot pdb> \
ligdir=<directory containing ligand sdf/mol2>
mapsdir=<directory containing SILCS FragMaps>
paramsfile=<params_custom.tmpl>
totruns=<# of runs>
The full list of user-definable parameters for params_custom.tmpl is:
CGENFF_RULES <cgenff rules_file>(required)This file is needed by the internal CGenFF library to determine the correct force-field parameters for the ligand. The default value is
${SILCSBIODIR}/data/cgenff/cgenff.rulesCGENFF_PAR <cgenff parameter file>(required)Along with the CGENFF_RULES file, this file is needed by the internal CGenFF library to determine the correct force-field parameters for the ligand. The default value is
${SILCSBIODIR}/data/cgenff/par_all36_cgenff.prmSILCS_RULES <silcs rules file>(required)This rules file is used to map the different atoms in the ligand to the corresponding SILCS FragMap types. This mapping is used to determine the appropriate “field” that will be applied to the different atoms in the ligand when attempting an MC-move. The default value is
${SILCSBIODIR}/data/silcs/silcs_classification_rules_2021_generic_apolar_scale_1e.datWhen
silcs_classification_rules_2021_generic_apolar_scale_1e.datis used, ligand atoms are assigned using generic classifications for mapping back to the FragMaps.Additional rules files are available in the
${SILCSBIODIR}/data/silcs/for other mapping schemes, including for more specific classifications and for using halogen FragMaps (see SILCS simulation setup with halogen probes).GFE_CAP <default: 3.0>Maximum allowable unfavorable GFE (kcal/mol) in the MC calculation.
RDIE <default: true>When true, the distance dependent dielectric (RDIE) scheme is used to treat intramolecular electrostatics. When false, CDIE (constant dielectric scheme) is used.
DIELEC_CONST <default: 4>Dielectric constant used in the intramolecular electrostatic interactions calculations.
# MINIMIZE_INPUT <default: false>
Perform minimization of input structure.
# MINIMIZE_BFGS <default: false>
Perform minimization of input structure using BFGS algorithm.
MIN_STEPS <default: 10000>Maximum number of steps of minimization performed using the steepest-descent algorithm with the ligand, before initiating MC simulation.
EMTOL <default: 0.01>Minimization is converged when the diff in total energy (totE) across the last 10 steps is smaller than this value. Once this criteria is satisfied minimization terminates.
MC_MOVE_RANGE <default:1.0 180.0 180.0>.Maximum range of translation, rigid body rotation and dihedral rotation per step of MC simulation.
MC_PRNT_FRQ <default: 0>Number of intermediate steps of MC to be written into OUTMCPDBFILE.
MC_STEPS <default: 10000>Number of steps of MC simulation to be performed per cycle.
SIM_ANNEAL_MOVE_RANGE <default:0.2 9.0 9.0>Maximum range of translation, rigid body rotation and dihedral rotation per step of simulated annealing simulation after MC simulation.
SIM_ANNEAL_STEPS <default: 40000>Number of steps of simulated annealing to be performed per cycle.
INIT_RUNS <default: 50>Number of MC/SA cycles before initiating checks for convergence.
NUM_TOL <default: 3>Number of top-scoring cycles with differences in LGFE less than DELTAE_TOLERANCE, before this simulation (run) is considered converged.
DELTAE_TOLERANCE <default: 0.5>When differences in LGFE of NUM_TOL most-favorable cycles are less than this defined tolerance value, convergence is reached and the program exits
DELTAE_BUFF <default: 10>Progression of MC+SA from one cycle to next is such that LGFE (of lowest conf) from MC should be less than (prev_min+deltae_buff). This ensures that N cycles are proceeding towards a minimum lower than that previously discovered lowest energy conformation.
TOTE_CRITERIA <default: false>When true, instead of LGFE, total energy (totE) of the system is used for convergence checks. Useful when running vacuum-phase MC simulations of the ligand.
TOT_RUNS <default: 250>Maximum number of MC simulation cycles. The program terminates if the
DELTAE_TOLERANCEcriteria is satisfied before reachingTOT_RUNS. Alternately, even if theDELTAE_TOLERANCEcriteria is not satisfied when the number of cycles executed reachesTOT_RUNS, the program terminates.RANDOM_SEED: <default: system-time>Seed used in MC simulation. When not set, system-time is used as a seed.
SIMULATION_CENTER: <x,y,z>Cartesian coordinates of where the MC simulation should be performed.
SIMULATION_RADIUS: <default: 10.0 A>Radius of the sphere within which MC simulation will be performed.
RANDOM_INIT_ORIENT: <true/false>When set to
TRUE,SIMULATION_CENTERshould also be set. The ligand is placed within a sphere of sizeSIMULATION_RADIUSin a random orientation and a random conformation.When set to
FALSE, then the center-of-geometry of the ligand is used as the center for the MC simulation. This is useful when the ligand pose in the pocket is well known.ATOM_TO_RESTRAIN: <atom number in sdf/mol2>When set, a spherical potential is applied to restrain the defined atom within the sphere during MC moves. This enables geometrically restraining a particular pharmacophore feature. Note, when using this feature, supply the full molecule with explicit hydrogens already added. Rrandom pocket pose and placement using
RANDOM_INIT_ORIENTtrue is incompatible with this option.When not set, the entire molecule is free to rotate/move/translate
ATOM_RESTRAINT_CENTER: <x,y,z>To be used in conjunction with
ATOM_TO_RESTRAINoption. This value is used to defined the center of the spherical potential.ATOM_RESTRAINT_RADIUS: <default: 1.0 A>To be used in conjunction with
ATOM_TO_RESTRAINoption. This value is used to defined the radius of the spherical potential. When not defined, then a default of 1 A is used.OUTRMSDFILE <output RMSD file>This file stores the RMSD and LGFE of the lowest energy conformation from each run of the MC/SA simulation. To be used in conjunction with
RANDOM_INIT_ORIENTset totrue.SILCSMAP <MapType> <map name> <scaling factor>(required)Multiple
SILCSMAPentries are typically defined, with each entry pointing to a FragMap filename<map name>.<scaling factor>is used to scale atomic Grid Free Energies (GFEs) for<MapType>atoms in the ligand being scored.<MapType>entries here must correspond to those defined in theSILCS_RULESrules file described above.OUTPUT_FORMAT [SDF|PDB](required)Choice of output format. SDF or PDB is supported.
OUTPUT_FILE <output file name>(required)This file stores the lowest energy conformation from each cycle of the MC/SA simulation.
LOGFILE <output log file>This file stores the energy statistics of the lowest energy conformation from each cycle of the MC/SA simulation.