SILCS-MC: Docking and Pose Refinement
SILCS-MC Background
The power of SILCS lies in the ability to use FragMaps to rapidly evaluate binding of diverse ligands to a target. SILCS-MC is Monte-Carlo (MC) sampling of ligands in translational, rotational, and torsional space in the field of the FragMaps and Exclusion Map. MC sampling uses CGenFF force field intramolecular energies, the Ligand Grid Free Energy (LGFE), which is the sum of atomic Grid Free Energies (GFEs), and the Exclusion Map. The Exclusion Map prevents ligand sampling where no probe or water molecules visited during SILCS simulations. SILCS-MC allows for rapid conformational sampling of the ligand while accounting for protein flexibility in a mean-field-like fashion since ligand affinity and volume exclusion are embedded in the combination of FragMaps and the Exclusion Map. Final ligand scoring is based on the LGFE score [14] [15]. In addition, the atomic GFE scores that comprise the LGFE score represent the free energy contribution of individual atoms to binding. This information can greatly facilitate the interpretation of experimental data and ligand design.
SILCS-MC can be used to generate and score binding poses for a ligand to a target using SILCS FragMaps that have been previously computed for that target (see SILCS Simulations). This can readily be done for a single ligand or a database of ligands. Two default conformational sampling protocols are available, “docking” and “pose refinement,” as described in detail below. The docking protocol is useful when no information is available about the binding pose, as it entails extensive translational, rotational, and intramolecular conformational sampling. The pose refinement protocol is useful when a reasonable starting pose for the ligand is available, for example to re-score poses output by another high-throughput in silico screening tool. Instructions for developing custom SILCS-MC ligand conformational sampling protocols are provided at the end of this chapter.
SILCS-MC Docking
Docking is exhaustive sampling of a ligand’s conformation in a given pocket to determine its most favorable orientation and internal geometry as defined by LGFE scoring. The pocket for the purposes of ligand sampling is defined as a sphere with a given radius and center; the same pocket is used for all ligands in a given SILCS-MC run. This protocol entails one MC run with the ligand for GPU jobs and five independent MC runs with the ligand for CPU jobs.
This protocol is recommended for ligands with diverse chemotypes and unknown binding poses. When the pose of a parent ligand is known and SILCS-MC evaluations are to be performed over a congeneric series, the pose refinement protocol is recommended instead (see below).
SILCS-MC Docking Using the SilcsBio GUI
Begin a new SILCS-MC Docking project:
Select New SILCS-MC Docking project from the Home page.
Enter a project name, select the remote server, and input files:
Enter a project name and select the remote server where the SILCS-MC docking jobs will run. Also, provide FragMap and protein input files. You may choose these files from the computer where you are running the SilcsBio GUI (“localhost”) or from any server you have previously configured, as described in File and Directory Selection. You will additionally need to provide a “Ligand SDF File” that contains the database of ligands to be docked. Note that users can modify ligands using the “Modify Ligand” utility available on the “Home Page” (see Modify Ligand for SILCS-MC).
Warning
Ligands in the “Ligand SDF File” must include all hydrogens, including pH-appropriate (de)protonations, and must have reasonable three dimensional conformations.

Select the SILCS-MC sampling protocol:
Once all information is entered correctly, press the “Setup” button at the bottom of the page. The page will update to list the number of ligands and show options for the sampling protocol (“Docking”, “Pose Refinement”, or “Pose Evaluation”) and the sampling region. Select the “Docking” option and then press the “Select Pocket” button.

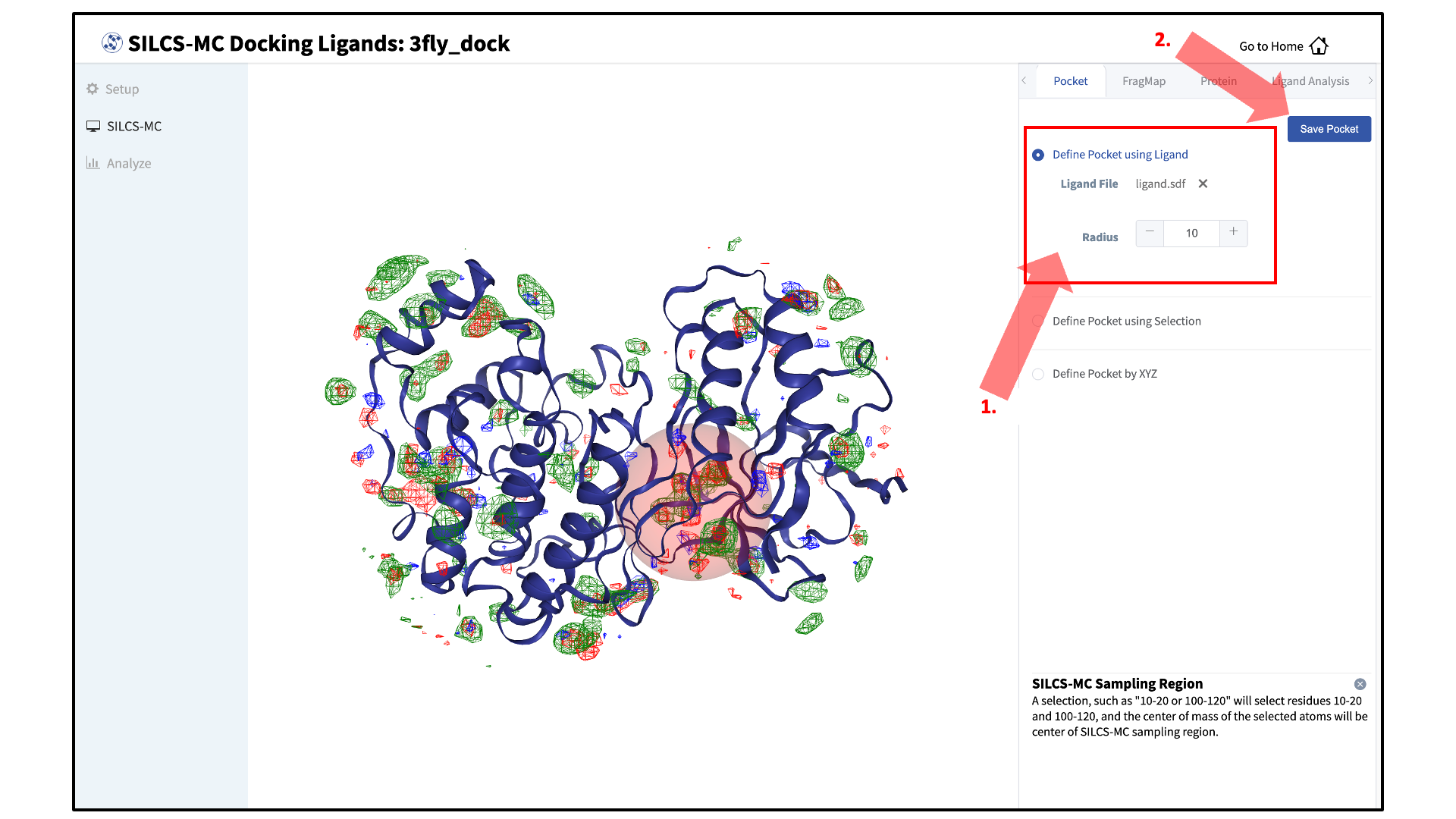
Define a SILCS-MC sampling region:
The GUI will now be showing the protein molecular graphic in the center pane. On the right-hand side in the “Pocket” tab, you can define the pocket center based on the center-of-geometry of a ligand pose (“Define Pocket using Ligand”), or a target residue selection (“Define Pocket using Selection”), or by directly entering an x, y, z coordinate (“Define Pocket by XYZ”). You may also adjust the sampling region’s radius (default value “10” Å) by using the “+/–” buttons. If it is difficult to see the spherical pocket definition in the center panel, hide the protein surface representation. Click on the “Save Pocket” button and the “OK” acknowledgement to continue.
Warning
Adjusting the radius of the spherical pocket may negatively impact the SILCS-MC sampling. Using a radius smaller than the default 10 Å reduces the size of the sampling region, which may disallow the generation of sufficient number of conformations leading to the optimal binding pose being missed. Using a radius larger than the default 10 Å increases the size of the sampling region, thereby increasing the computational cost and time of the SILCS-MC docking runs and increasing the number of irrelevant binding poses.
Launch SILCS-MC jobs:
You will be returned to the previous screen, which now includes “Sampling Region” information consisting of the spherical pocket center and radius.
Tip
If you ran Halogen SILCS simulations for your target, you can include the Halogen SILCS FragMaps in the SILCS-MC posing and scoring by checking the “Include Halogen FragMaps” box.

You may now click the “Run SILCS-MC” button to start the SILCS-MC docking. Doing so will submit jobs to the remote server job schedular and list them in a pop-over window.
Once all jobs are submitted, you may click on the “Setup Successful” button to dismiss the pop-over window and return to the previous screen, which will now show a “Simulation Progress” section. You may update this section by scrolling to the bottom of the screen and clicking the “Refresh” button. This will update the progress bars for all of the ligands being docked.
Visualize SILCS-MC docked poses:
Once progress bars for all ligands reach 100%, click on the “Show Chart” button above the “Simulation Progress” section to proceed.

Upon successful completion of this command in the pop-over window, you may click on the green “Data collection finished” button to return to the GUI. A new tab, labeled “Chart” will have been created in the right-hand panel. Under that tab, clicking on a ligand name will display that ligand.

Choosing the “GFE” tab in the right-hand panel will allow you to click on individual atoms within a ligand to see their atomic GFE values. The “GFE” tab will also display the sum of all GFE values of atoms selected in that ligand.

SILCS-MC Docking Using the CLI
Ligands can be docked using SILCS-MC docking through the following steps:
Launch SILCS-MC docking runs:
To set up and run SILCS-MC docking from the command line interface, create a directory containing all the ligands to be evaluated. Each ligand can be stored as a separate SDF or Mol2 file. Alternatively, all the ligands can be combined into a single SD file. With this information, enter the following command to set up and launch SILCS-MC docking runs:
${SILCSBIODIR}/silcs-mc/1_run_silcsmc_exhaustive prot=<prot pdb> ligdir=<ligand mol2/sdf directory>Required parameters:
Path and name of protein PDB file:
prot=<protein pdb file>
Path and name of directory containing ligand SDF or Mol2 files:
ligdir=<location and name of directory containing ligand mol2/sdf>
If the ligdir parameter is used, only one molecule per file under
ligdirwill be processed for SILCS-MC docking. Subdirectories underligdirwill also be processed. For an SD/SDF file containing multiple molecules, usesdfile=<path to sdfile>instead of theligdirparameter.Warning
Ligands, regardless of file format, must include all hydrogens, including pH-appropriate (de)protonations, and must have reasonable three dimensional conformations.
Optional parameters:
Path and name of directory containing FragMaps:
mapsdir=<location and name of directory containing FragMaps; default=maps>
Name of the directory containing the SILCS-MC output:
silcsmcdir=<name of output directory; default=3_silcsmc>
Center of the spherical sampling region:
center=<"x,y,z"; e.g., center="20,30,-2"; default=ligand's center of mass>
If the center is not specified by the user, the default center will be calculated from the input ligand’s center of mass, and the initial conformation of the ligand in the SILCS-MC docking runs will correspond to the conformation of the ligand in the input Mol2 or SD file. If the center is specified, then the conformation and orientation of the ligand will be randomized prior to the SILCS-MC docking runs.
Radius of the spherical sampling region:
radius=<SILCS-MC calculation radius from center in Å; default=10.0>
Path and name of ligand SD file:
sdfile=<location and name of SD file, this option will overwrite ligdir>
If the
sdfileoption is used, theligdiroption is not needed and any input for theligdiroption will be overwritten. Thesdfileoption is recommended if the user has an SD file containing all ligands under investigation.Alignment to reference molecule prior to SILCS-MC docking:
refsdf=<location and name of SD file of reference molecule to align to; default=None>
If the
refsdfoption is used, the ligands insdfilewill be aligned to the reference molecule before the SILCS-MC docking runs are initiated. The alignment is performed using the Open3DAlign(O3A) algorithm [16] available in RDKit. Currently, this option is only available using thesdfileoption, and is not available using theligdiroption.Inclusion of halogen FragMaps:
halogen=<use halogen maps; true/false; default=false>
If halogen FragMaps have been generated (see Setup with halogen probes), they can be included in the SILCS-MC calculation to improve the scoring of halogen-containing compounds. To do so, add the
halogen=trueoption.Atom classification scheme (ACS):
class=<atom classification scheme (generic or specific); default=generic>
Atomic GFEs are assigned based on the atom’s classification and its overlap with the corresponding FragMap. The atom’s classification is assigned based on the ACS used. Users can choose between generic and specific ACS using the
classoption. The default ACS is thegenericACS in which the generic apolar, donor, and acceptor maps are used to score corresponding atoms. In thespecificACS, specific maps (e.g., formamide nitrogen FragMaps and imidizole donor nitrogen FragMaps are kept separate and are not grouped into generic donor FragMaps) to score corresponding atoms. The schemes are stored in${SILCSBIODIR}/data/silcs/. Further details regarding ACS and how they affect atomic GFEs and LGFEs are provided in ref. [17].Option to bundle jobs:
bundle=<bundle multiple (single) jobs into larger jobs; true/false; default=false>
When
bundle=truemultiple (single) jobs will be bundled into a single larger job. The number of jobs to be bundled can be set with thenprocparameter.Number of jobs to bundle (when
bundle=true):nproc=<number of jobs to bundle; default=8>
By default number of jobs to bundle is determined by the number of CPU cores on the compute nodes available to the user. The user can override this using the
nprocparameter.Path to python executable:
python=<path to python executable; default=$(which python)>
The default python path will be checked using the
which pythoncommand.Use GPU version of SILCS-MC:
gpu=<use GPU; true/false; default=true>
By default, the GPU version of SILCS-MC is used. To use the CPU version, use
gpu=false. The GPU version of SILCS-MC is only available on computers with NVIDIA GPUs and CUDA version 11 or later. The CPU version of SILCS-MC is available on all machines, however it is significantly slower than the GPU version. When using the CPU version of SILCS-MC, the SILCS-MC calculation will be split into five independent, single-core serial jobs per ligand. Also note that thenprocparameter must not be changed from its default value when using thegpu=false bundle=trueoptions to bundle CPU jobs.
SILCS-MC Docking with multiple ligands:
Users may dock multiple ligands to a binding site simultaneously. To dock multiple ligands simultaneously, the user can either provide an additional directory containing all of the additional ligands to be docked as separate files in Mol2 or SD file format or an additional single SD file containing all of the ligands to be docked. These additional ligands, herein referred to as secondary ligands, are docked in the same pocket as the primary ligand. To perform SILCS-MC docking with multiple ligands, the user can set the following parameters to dock the primary ligand with secondary ligands simultaneously:
Secondary Ligand Parameters:
Perform SILCS-MC docking with secondary ligands:
seclig=<true/false; default=false>
By default, the SILCS-MC docking will be performed with the primary ligand only. Users can perform SILCS-MC docking on multiple ligands simultaneously by setting
seclig=true. When set to true, SILCS-MC will be performed on both the primary ligand (specified byligdirorsdfile) and the secondary ligand. By default, 15 water molecules will be used as secondary ligands, however, the secondary ligand may also be specified withsecligdirorsecsdfile(described below).Path and name of directory containing secondary ligand SD or Mol2 files:
secligdir=<path to directory containing secondary ligand mol2/sdf files>
When
seclig=true, by default thesecligdiroption is internally set to${SILCSBIODIR}/templates/silcs-mc/secligdir, which contains 15 water molecules. Users can provide a different set of secondary ligands by setting thesecligdiroption to the path of the directory containing the desired secondary ligand Mol2 or SD files.Tip
To increase or decrease the number of water molecules simultaneously docked with the primary ligand, the user can copy the desired number of water molecules from the default
secligdirdirectory (${SILCSBIODIR}/templates/silcs-mc/secligdir) to a new directory and set thesecligdiroption to the new directory. It does not matter if the coordinates of two water molecules are the same, as long as the file names are different.Path and name of secondary ligand SD file:
secsdfile=<path to sdf file of the secondary ligands; this option will overwrite secligdir>
If the
secsdfileoption is used, then thesecligdiroption is not needed and any input used for thesecligdiroption will be overwritten. Thesecsdfileoption is recommended if the user has an SD file containing all secondary ligands to be used.Radius of the spherical sampling region for secondary ligands:
secradius=<MC-SILCS calculation radius from center for secondary ligands; default=10.0>
When
seclig=true, the default radius for the secondary ligands is set to 10.0 Å. Based on the size of the primary and secondary ligands, the user can adjust thesecradiusvalue to ensure that the secondary ligands are sampled around the longest axis of the primary ligand. The center of the spherical sampling region for the secondary ligands is the same as the geometric center of the primary ligand.
Evaluate docked poses:
The SILCS-MC tool set allows users to easily evaluate docked poses generated from the SILCS-MC docking runs. Users can rank order and collect the docked poses using
2_calc_lgfe_min_avg_sd(see Best-Pose Retrieval) and extract SILCS simulation snapshots in which the protein conformation best complements the docked conformation and orientation of the ligand using3_scan_traj(see Best Protein–Ligand Complex Retrieval).For details on evaluating the resulting SILCS-MC docked poses, please refer to Assessment of SILCS-MC Docked/Refined Poses in CLI.
SILCS-MC Docking Protocol Details
On computers using NVIDIA GPU with CUDA version 11 or later, SILCS-MC Docking spawns
one independent job on one GPU and one CPU per ligand. On computers without NVIDIA GPU,
SILCS-MC Docking spawns five independent single-core serial jobs per ligand.
Each SILCS-MC run involves 1250 cycles (on computers with NVIDIA GPU) or a maximum of 250
cycles and a minimum of 50 cycles
(on computers without NVIDIA GPU) of MC/SA sampling of the ligand within a
defined spherical sampling space.
Each of these cycles,
regardless of GPU or CPU usage, consists of 10,000 steps of MC at a high temperature
followed by 40,000 steps of SA towards a lower temperature. At the beginning of
each cycle, the ligand will be reoriented within the predefined sphere.
The MC sampling has three types of moves: i) molecular translations with a
maximum step size of 1 Å, ii) molecular rotation with a maximum step size of
180 degrees, and iii) intramolecular dihedral rotations with a maximum
step size of 180 degrees. For intramolecular dihedral rotations, only the
rotatable dihedral angles are selected for MC moves. The lowest LGFE scoring
pose from the MC sampling is used as starting pose in the following SA
sampling. The SA sampling also involves the same three types of moves, but
with a smaller step size compared to the MC sampling: i) molecular
translations with a maximum step size of 0.2 Å, ii) molecular rotation with a
maximum step size of 9 degrees and, iii) intramolecular dihedral rotations with
a maximum step size of 9 degrees. The lowest LGFE scoring pose from the SA is
saved in a multi-frame SD file: 3_silcsmc/<run>/out/<lig>.sdf.
On computers using NVIDIA GPU, the MC/SA procedure continues until 1250 cycles have been completed. On computers without GPU, the LGFE score difference between the top three poses (defined by lowest LGFE scores) are evaluated after a minimum of 50 MC/SA cycles. If the LGFE score difference between the 3 poses is less than 0.5 kcal/mol, then that run is considered converged and terminated. If the top three scored poses are separated by more 0.5 than kcal/mol, the MC/SA procedure continues either until the convergence criterion is met or until a maximum of 250 MC/SA cycles have been completed with the most favorable LGFE pose selected.
SILCS-MC Docking with Secondary Ligands (Water Molecules)
When including secondary ligands, the SILCS-MC docking protocol extends the MC steps to include the secondary ligands in addition to the primary ligand. The MC step sizes for the secondary ligands (by default water molecules) are set to 0.5 Å for translation, 180 degrees for rotation, and 180 degrees for dihedral rotation. The SA step sizes for the secondary ligands are set to 0.2 Å for translation, 9 degrees for rotation, and 9 degrees for dihedral rotation. The sampling region for the secondary ligands is set to a sphere centered at the geometric center of the primary ligand with a 10 Å (default) radius. The inclusion of water molecular as secondary ligands in SILCS-MC docking has been validated against crystal structures of protein–ligand complexes containing water mediated interactions [18].
SILCS-MC Pose Refinement
The pose refinement protocol is designed to limit conformational sampling near the ligand input pose supplied by the user. Pose refinement is appropriate when there is high confidence in the input parent ligand pose. The sphere center for the pocket definition is automatically and independently computed for each and every ligand and is the center-of-geometry of that ligand’s input coordinates.
SILCS-MC Pose Refinement Using the SilcsBio GUI
To use SILCS-MC Pose Refinement in the SilcsBio GUI, please see the previous section on
running SILCS-MC docking from the SilcsBio GUI (SILCS-MC Docking Using the SilcsBio GUI); in
Step 3, select the “Pose Refinement” option. Note that there
will be no “Select Pocket” step, as pose refinement assigns the pocket center
based on the center-of-geometry of the input ligand on a per-ligand basis. For multiple
input ligands in a single .sdf file, each ligand will have its own
center-of-geometry used to define the pocket center.
SILCS-MC Pose Refinement Using the CLI
Launch SILCS-MC pose refinement runs:
To set up and run SILCS-MC pose refinement, create a directory containing all the ligands to be evaluated. Each ligand can be stored as a separate
.mol2or.sdffile. Alternatively, all the ligands can be combined into a single.sdffile. With this information, enter the following command to set up and launch SILCS-MC refinement runs:${SILCSBIODIR}/silcs-mc/1_run_silcsmc_local prot=<prot pdb> ligdir=<directory containing ligand mol2/sdf>Required parameters:
Path and name of protein PDB file:
prot=<protein pdb file>
Path and name of directory containing ligand SDF or Mol2 files:
ligdir=<location and name of directory containing ligand mol2/sdf>
If the ligdir option is used, only one molecule per file under
ligdirwill be processed for SILCS-MC docking. Subdirectories underligdirwill also be processed. For an SD/SDF file containing multiple molecules, usesdfile=<path to sdfile>instead of theligdiroption.Warning
Ligands, regardless of file format, must include all hydrogens, including pH-appropriate (de)protonations, and must have reasonable three dimensional conformations.
Optional parameters:
Path and name of directory containing FragMaps:
mapsdir=<location and name of directory containing FragMaps; default=maps>
Name of the directory containing the SILCS-MC output:
silcsmcdir=<name of output directory; default=3_silcsmc>
Path and name of ligand SD file:
sdfile=<location and name of SD file, this option will overwrite ligdir>
If the
sdfileoption is used, theligdiroption is not needed and any input for theligdiroption will be overwritten. Thesdfileoption is recommended if the user has an SD file containing all ligands under investigation.Alignment to reference molecule prior to SILCS-MC docking:
refsdf=<location and name of SD file of reference molecule to align to; default=None>
If the
refsdfoption is used, the ligands insdfilewill be aligned to the reference molecule before the SILCS-MC docking runs are initiated. The alignment is performed using the Open3DAlign(O3A) algorithm [16] available in RDKit. Currently, this option is only available using thesdfileoption, and is not available using theligdiroption.Inclusion of halogen FragMaps:
halogen=<use halogen maps; true/false; default=false>
If halogen FragMaps have been generated (see Setup with halogen probes), they can be included in the SILCS-MC calculation to improve the scoring of halogen-containing compounds. To do so, add the
halogen=trueoption.Atom classification scheme (ACS):
class=<atom classification scheme (generic or specific); default=generic>
Atomic GFEs are assigned based on the atom’s classification and its overlap with the corresponding FragMap. The atom’s classification is assigned based on the ACS used. Users can choose between generic and specific ACS using the
classoption. The default ACS is thegenericACS in which the generic apolar, donor, and acceptor maps are used to score corresponding atoms. In thespecificACS, specific maps (e.g., formamide nitrogen FragMaps and imidizole donor nitrogen FragMaps are kept separate and are not grouped into generic donor FragMaps) to score corresponding atoms. The schemes are stored in${SILCSBIODIR}/data/silcs/. Further details regarding ACS and how they affect atomic GFEs and LGFEs are provided in ref. [17].Option to bundle jobs:
bundle=<bundle multiple (single) jobs into larger jobs; true/false; default=false>
When
bundle=truemultiple (single) jobs will be bundled into a single larger job. The number of jobs to be bundled can be set with thenprocparameter.Number of jobs to bundle (when
bundle=true):nproc=<number of jobs to bundle; default=8>
By default number of jobs to bundle is determined by the number of CPU cores on the compute nodes available to the user. The user can override this by setting the
nprocparameter.Path to python executable:
python=path to python executable; default=$(which python)>
The default python path will be checked using the
which pythoncommand.Use GPU version of SILCS-MC:
gpu=<use GPU; true/false; default=true>
By default, the GPU version of SILCS-MC is used. To use the CPU version, use
gpu=false. The GPU version of SILCS-MC is only available on computers with NVIDIA GPUs and CUDA version 11 or later. The CPU version of SILCS-MC is available on all machines, however it is significantly slower than the GPU version. When using the CPU version of SILCS-MC, the SILCS-MC calculation will be split into five independent, single-core serial jobs per ligand. Also note that thenprocparameter must not be changed from its default value when using thegpu=false bundle=trueoptions to bundle CPU jobs.
Secondary Ligand Parameters:
Perform SILCS-MC Pose Refinement with secondary ligands:
seclig=<true/false; default=false>
By default, the SILCS-MC pose refinement will be performed with the primary ligand only. Users can perform SILCS-MC pose refinement on the primary ligand with SILCS-MC docking on multiple ligands simultaneously by setting
seclig=true. When set to true, SILCS-MC pose refinment will be performed on the primary ligand (specified byligdirorsdfile) and SILCS-MC docking will be performed on the secondary ligand. By default, 15 water molecules will be used as secondary ligands, however, the secondary ligand may also be specified withsecligdirorsecsdfile(described below).Path and name of directory containing secondary ligand SDF or Mol2 files:
secligdir=<path to directory containing secondary ligand mol2/sdf files>
When
seclig=true, by default thesecligdiroption is internally set to${SILCSBIODIR}/templates/silcs-mc/secligdir, which contains 15 water molecules. Users can provide a different set of secondary ligands by setting thesecligdiroption to the path of the directory containing the desired secondary ligand Mol2 or SD files.Tip
To increase or decrease the number of water molecules simultaneously docked with the primary ligand, the user can copy the desired number of water molecules from the default
secligdirdirectory (${SILCSBIODIR}/templates/silcs-mc/secligdir) to a new directory and set thesecligdiroption to the new directory. It does not matter if the coordinates of two water molecules are the same, as long as the file names are different.Path and name of secondary ligand SD file:
secsdfile=<path to sdf file of the secondary ligands; this option will overwrite secligdir>
If the
secsdfileoption is used, thesecligdiroption is not needed, and any input for thesecligdiroption will be overwritten. Thesecsdfileoption is recommended if the user has an SDF file containing all secondary ligands to be used.
Evaluate refined poses:
The SILCS-MC tool set allows users to easily evaluate refined poses generated from the SILCS-MC refinement runs. Users can rank order and collect the resulting poses using
2_calc_lgfe_min_avg_sd(see Best-Pose Retrieval) and extract SILCS simulation snapshots in which the protein conformation best complements the refined conformation and orientation of the ligand using3_scan_traj(see Best Protein–Ligand Complex Retrieval).For details on evaluating the resulting SILCS-MC refined poses, please refer to Assessment of SILCS-MC Docked/Refined Poses in CLI.
SILCS-MC Pose Refinement Protocol Details
On computers using NVIDIA GPU with CUDA version 11 or later, SILCS-MC Pose Refinement
spawns one independent job on one GPU and one CPU per ligand. On computers without
NVIDIA GPU, SILCS-MC Pose Refinement spawns one independent single-core serial jobs
per ligand.
Each SILCS-MC run involves 1250 cycles (on computers with NVIDIA GPU) or 50 cycles
(on computers without NVIDIA GPU) of MC/SA sampling of the ligand within a 1 Å sphere.
The center of the sphere is defined
as the center-of-geometry of the input ligand pose.
Each of these cycles, regardless of GPU or CPU usage,
consists of 100 steps of MC at high temperature followed by 1000 steps
of SA towards a lower temperature. At the beginning of each cycle, the
ligand orientation/conformation will be reset to the one found in the
input file. MC sampling moves are: i) molecular translation with a
maximum step size of 0.5 Å, ii) molecular rotation with a maximum step
size of 15 degrees, and iii) intramolecular dihedral rotation with a
maximum step size of 180 degrees. For intramolecular dihedral rotation,
only the rotatable dihedral angles are selected for MC moves. The lowest
LGFE scoring pose from the MC sampling is used as starting pose in the
following SA sampling. SA sampling moves are smaller than for the MC
phase: i) molecular translation with a maximum step size of 0.2 Å, ii)
molecular rotation with a maximum step size of 9 degrees, iii)
intramolecular dihedral rotation with a maximum step size of 9 degrees.
The lowest LGFE scoring pose from the SA is saved in the multi-frame SD
file 3_silcsmc/<run>/out/<lig>.sdf.
On computers using NVIDIA GPU, the MC/SA procedure continues until 1250 cycles have been completed. On computers without GPU, the LGFE score difference between the top three poses (defined by lowest LGFE scores) are evaluated after a minimum of 50 MC/SA cycles. If the LGFE score difference between the 3 poses is less than 0.5 kcal/mol, then that run is considered converged and terminated. If the top three scored poses are separated by more 0.5 than kcal/mol, the MC/SA procedure continues either until the convergence criterion is met or until a maximum of 250 MC/SA cycles have been completed with the most favorable LGFE pose selected.
For secondary ligands, the protocol is same as in the SILCS-MC docking protocol as described in SILCS-MC Docking with Secondary Ligands (Water Molecules).
SILCS-MC Pose Evaluation
SILCS-MC pose evaluation allows the user to evaluate the LGFE of a crystal ligand or a ligand already docked to a target protein for which FragMaps are available. The SILCS-MC pose evaluation protocol evaluates the LGFE score based on the input pose of the ligand. No conformational sampling of the ligand is performed.
SILCS-MC Pose Evaluation Using the SilcsBio GUI
To calculate the LGFE of a pose without re-docking or refining the ligand conformation and orientation, please see the previous section on running SILCS-MC docking from the SilcsBio GUI (SILCS-MC Docking Using the SilcsBio GUI); in Step 3, select the “Pose Evaluation” option. Note that there will be no “Select Pocket” step, as pose evaluation will only calculate the LGFE of the input ligand in its original orientation and conformation.
SILCS-MC Pose Evaluation Using the CLI
Launch SILCS-MC pose evaluation runs:
To set up and run SILCS-MC pose evaluation, create a directory containing all the ligands to be evaluated. Each ligand can be stored as a separate
.mol2or.sdffile. Alternatively, all the ligands can be combined into a single.sdffile. With this information, enter the following command to set up and launch SILCS-MC refinement runs:${SILCSBIODIR}/silcs-mc/1_run_silcsmc_fixedpose prot=<prot pdb> ligdir=<directory containing ligand mol2/sdf>Required parameters:
Path and name of protein PDB file:
prot=<protein pdb file>
Path and name of directory containing ligand SDF or Mol2 files:
ligdir=<location and name of directory containing ligand mol2/sdf>
If the ligdir option is used, only one molecule per file under
ligdirwill be processed for SILCS-MC docking. Subdirectories underligdirwill also be processed. For an SD/SDF file containing multiple molecules, usesdfile=<path to sdfile>instead of theligdiroption.Warning
Ligands, regardless of file format, must include all hydrogens, including pH-appropriate (de)protonations, and must have reasonable three dimensional conformations.
Optional parameters:
Path and name of directory containing FragMaps:
mapsdir=<location and name of directory containing FragMaps; default=maps>
Name of the directory containing the SILCS-MC output:
silcsmcdir=<name of output directory; default=3_silcsmc>
Path and name of ligand SD file:
sdfile=<location and name of SD file, this option will overwrite ligdir>
If the
sdfileoption is used, theligdiroption is not needed, and any input for theligdiroption will be overwritten. Thesdfileoption is recommended if the user has an SD file containing all ligands under investigation.Inclusion of halogen FragMaps:
halogen=<use halogen maps; true/false; default=false>
If halogen FragMaps have been generated (see Setup with halogen probes), they can be included in the SILCS-MC calculation to improve the scoring of halogen-containing compounds. To do so, add the
halogen=trueoption to the1_run_silcsmc_exhaustivecommand.Atom classification scheme (ACS):
class=<atom classification scheme (generic or specific); default=generic>
Atomic GFEs are assigned based on the atom’s classification and its overlap with the corresponding FragMap. The atom’s classification is assigned based on the ACS used. Users can choose between generic and specific ACS using the
classoption. The default ACS is thegenericACS in which the generic apolar, donor, and acceptor maps are used to score corresponding atoms. In thespecificACS, specific maps (e.g., formamide nitrogen FragMaps and imidizole donor nitrogen FragMaps are kept separate and are not grouped into generic donor FragMaps) to score corresponding atoms. The schemes are stored in${SILCSBIODIR}/data/silcs/. Further details regarding ACS and how they affect atomic GFEs and LGFEs are provided in ref. [17].Option to bundle jobs:
bundle=<bundle multiple (single) jobs into larger jobs; true/false; default=false>
When
bundle=truemultiple (single) jobs will be bundled into a single, larger job. The number of jobs to be bundled can be set with thenprocparameter.Number of jobs to bundle (when
bundle=true):nproc=<number of jobs to bundle; default=8>
Path to python executable:
python=path to python executable; default=$(which python)>
The default python path will be checked using the
which pythoncommand.Use GPU version of SILCS-MC:
gpu=<use GPU; true/false; default=true>
By default, the GPU version of SILCS-MC is used. To use the CPU version, set
gpu=false. The GPU version of SILCS-MC is only available on computers with NVIDIA GPUs and CUDA version 11 or later. The CPU version of SILCS-MC is available on all machines, however it is significantly slower than the GPU version. When using the CPU version of SILCS-MC, the SILCS-MC calculation will be performed as a single independent, single-core serial job per ligand. Also note that thenprocparameter must not be changed from its default value when using thegpu=false bundle=trueoptions to bundle CPU jobs.
Secondary Ligand Parameters:
Perform SILCS-MC Pose Evaluation with secondary ligands:
seclig=<true/false; default=false>
By default, the SILCS-MC pose evaluation will be performed on the primary ligand only. Users can perform SILCS-MC pose evaluation on multiple ligands simultaneously by setting
seclig=true. When set to true, the user will have to provide a set of secondary ligands with either thesecligdirorsecsdfileoptions as described below.Path and name of directory containing secondary ligand SDF or Mol2 files:
secligdir=<path to directory containing secondary ligand mol2/sdf files>
Users can provide a set of secondary ligands by setting the
secligdiroption to the path of the directory containing the secondary ligand Mol2 or SD files.Path and name of secondary ligand SD file:
secsdfile=<path to sdf file of the secondary ligands; this option will overwrite secligdir>
If the
secsdfileoption is used, thesecligdiroption is not needed, and any input for thesecligdiroption will be overwritten. Thesecsdfileoption is recommended if the user has an SDF file containing all secondary ligands to be used.
Evaluate poses:
To extract the LGFE scores calculated by SILCS-MC pose evaluation, use
2_calc_lgfe_min_avg_sdfollowing the instructions detailed in Best-Pose Retrieval. Users can additionally extract SILCS simulation snapshots in which the protein conformation best complements the refined conformation and orientation of the ligand using3_scan_traj(see Best Protein–Ligand Complex Retrieval).
SILCS-MC Pose Evaluation Protocol Details
Pose evaluation initiates one single-GPU and single-CPU job (or single-core serial job on computers without NVIDIA GPU or without CUDA version 11 or later) per ligand, and involves a single step evaluation of LGFE. Technically, all step sizes are set to zero and only 1 step of MC and SA is performed in order to get the output LGFE. When including secondary ligands, all step sizes for the secondary ligands are also set to zero.
SILCS-MC Docking in Restrained Mode
SILCS-MC restrained docking allows the user to dock ligands that form covalent bonds with the target protein. SILCS-MC restrained docking involves exhaustive sampling of a ligand’s conformation in a given pocket to determine its most favorable orientation, as in SILCS-MC docking with the exception that the ligand sampling is restricted by a constraint on a user-defined reactive atom of the ligand. All other sampling parameters are the same as in SILCS-MC docking.
SILCS-MC Restrained Mode Docking Using the CLI
Launch SILCS-MC Restrained Mode docking runs:
To set up and run SILCS-MC Restrained Mode docking, create a directory containing all of the ligands to be evaluated. Each ligand can be stored as a separate Mol2 or SD file. Alternatively, all the ligands can be combined into a single SD file. In addition, a restraints file that specifies the atom index number of the atom to be restrained for each ligand is required. With this information, enter the following command to set up and launch SILCS-MC docking runs in Restrained Mode:
${SILCSBIODIR}/silcs-mc/1_run_silcsmc_restrained prot=<prot pdb> \ ligdir=<directory containing ligand mol2/sdf> \ rstrfile=<location and name of restraints file>Required parameters:
Path and name of protein PDB file:
prot=<protein pdb file>
Path and name of directory containing ligand SDF or Mol2 files:
ligdir=<location and name of directory containing ligand mol2/sdf>
If the ligdir option is used, only one molecule per file under
ligdirwill be processed for SILCS-MC docking. Subdirectories underligdirwill also be processed. For an SD/SDF file containing multiple molecules, usesdfile=<path to sdfile>instead of theligdiroption.Warning
Ligands, regardless of file format, must include all hydrogens, including pH-appropriate (de)protonations, and must have reasonable three dimensional conformations.
Tip
If using the SILCS-MC restrained mode docking to approximate the binding of covalently bonded inhibitors, then the chemical structure of the ligand in the covalent bond should be considered when preparing the ligand Mol2/SD file. The ligands being docked can be prepared with the reactive atom in the form after a covalent bond has been formed with the target protein. Hydrogen atoms could be temporarily added to the reactive atom (and adjacent atoms as needed) to facilitate the restrained docking. For example, the C=C double bond of an acrylamide molecule could be changed to a C-C single bond for a covalently bonded acrylamide moiety, where the constraint imposed on the ligand C atom involved in covalent bond being formed with, for example, the S of a Cysteine. Note that the ligand’s chemical structure must be a stable species.
Path and name of restraints file:
rstrfile=<location and name of restraints file>
The restraints file informs the program which atom(s) in the ligand are restrained in the docking runs. The restraints file should contain the following information:
<ligand_name> <atom_index> <x> <y> <z> <radius>where,
ligand_name: name of the ligand in the Mol2/SD file (not the file name)atom_index: index of the atom to be constrainedx, y, z: coordinates for the center of the constraint in Angstromsradius: radius of the constraint in Angstroms (Recommended: 0.5 - 1.5)
Optional parameters:
Path and name of directory containing FragMaps:
mapsdir=<location and name of directory containing FragMaps; default=maps>
Name of the directory containing the SILCS-MC output:
silcsmcdir=<name of output directory; default=3_silcsmc>
Path and name of ligand SD file:
sdfile=<location and name of SD file; this option will overwrite ligdir>
If the
sdfileoption is used, theligdiroption is not needed and any input for theligdiroption will be overwritten. Thesdfileoption is recommended if the user has an SD file containing all ligands under investigation.Alignment to reference molecule prior to SILCS-MC docking:
refsdf=<location and name of SD file of reference molecule to align to; default=None>
If the
refsdfoption is used, the ligands insdfilewill be aligned to the reference molecule before the SILCS-MC docking runs are initiated. The alignment is performed using the Open3DAlign(O3A) algorithm [16] available in RDKit. Currently, this option is only available using thesdfileoption, and is not available using theligdiroption.Inclusion of halogen FragMaps:
halogen=<use halogen maps; true/false; default=false>
If halogen FragMaps have been generated (see Setup with halogen probes), they can be included in the SILCS-MC calculation to improve the scoring of halogen-containing compounds. To do so, add the
halogen=trueoption to the1_run_silcsmc_exhaustivecommand.Atom classification scheme:
class=<atom classification scheme (generic or specific); default=generic>
Users can use custom atom classification schemes if they wish.
Option to bundle jobs:
bundle=<bundle multiple (single) jobs into larger jobs; true/false; default=false>
When
bundle=truemultiple (single) jobs will be bundled into a single larger job. The number of jobs to be bundled can be set with thenprocparameter.Number of jobs to bundle (when
bundle=true):nproc=<number of jobs to bundle; default=8>
Path to python executable:
python=path to python executable; default=$(which python)>
The default python path will be checked using the
which pythoncommand.
Additional Parameters
Center of the spherical sampling region:
center=<"x,y,z"; e.g., center="20,30,-2"; default=ligand's center of mass>
If the center is not specified by the user, the default center will be calculated from the input ligand’s center of mass, and the initial conformation of the ligand in the SILCS-MC docking runs will correspond to the conformation of the ligand in the input Mol2 or SD file. If the center is specified, then the conformation and orientation of the ligand will be randomized prior to the SILCS-MC docking runs.
Radius of the spherical sampling region:
radius=<radius from center in Å; default=10.0>
Evaluate docked poses:
The SILCS-MC tool set allows users to easily evaluate docked poses generated from the SILCS-MC docking runs. Users can rank order and collect the docked poses using
2_calc_lgfe_min_avg_sd(see Best-Pose Retrieval) and extract SILCS simulation snapshots in which the protein conformation best complements the docked conformation and orientation of the ligand using3_scan_traj(see Best Protein–Ligand Complex Retrieval).For details on evaluating the resulting SILCS-MC docked poses, please refer to Assessment of SILCS-MC Docked/Refined Poses in CLI.
SILCS-MC Restrained Mode Docking Protocol Details
The SILCS-MC Restrained Mode docking protocol is similar to the SILCS-MC docking protocol, but with the addition of a contraint on user-specified specified atoms of the ligand. The atom(s) are specified by the user in the restraints file. Additionally, the restraints file should contain the center and the radius of the contraint. The radius of the constraint is the distance from the center within which the reactive atom is allowed to move. A radius of 0.5 - 1.5 Å is recommended.
SILCS-MC Docking in Slab Mode
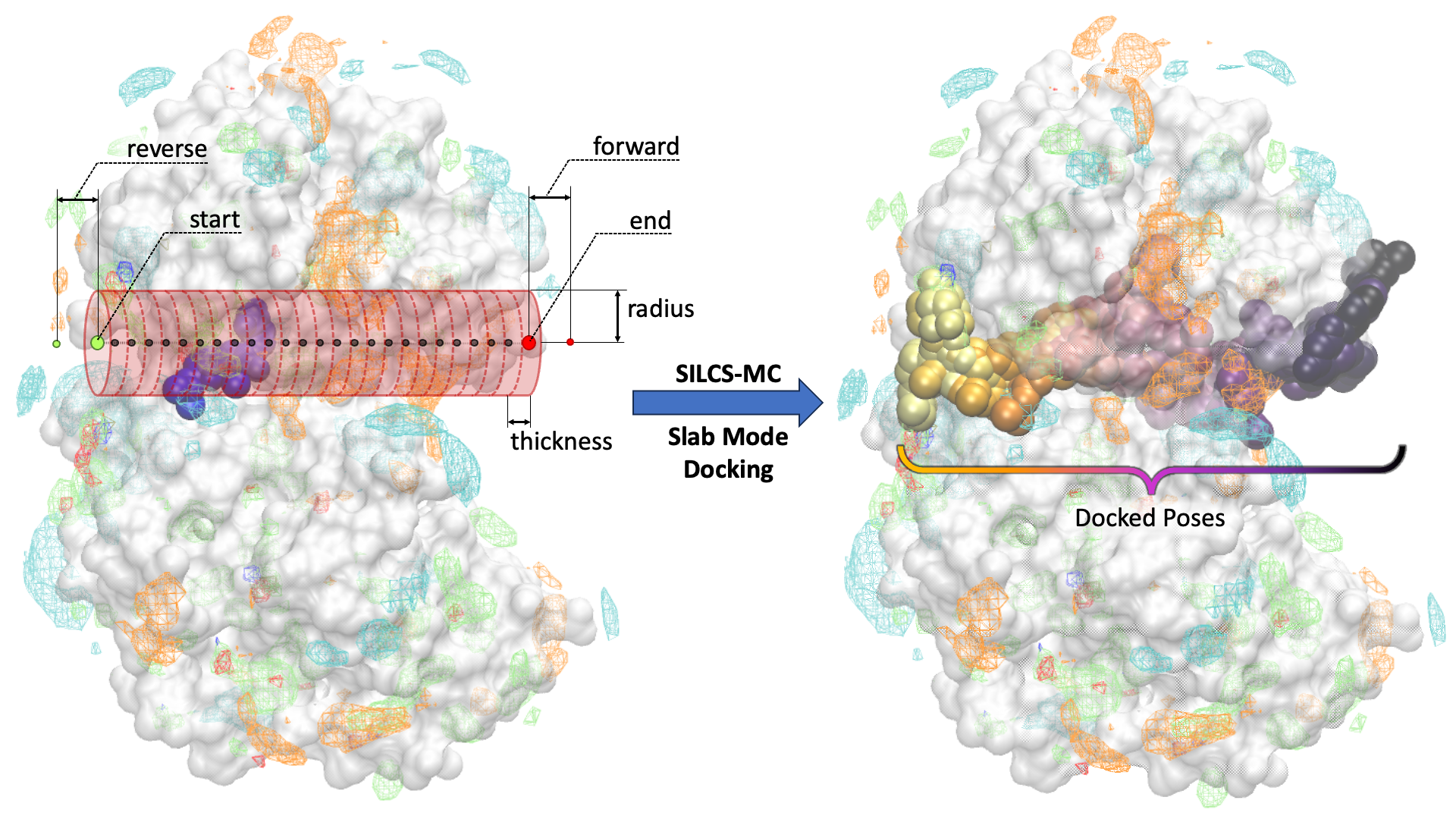
For a ligand to reach the active site of a protein, the ligand must diffuse into the active site. Characterizing ligand binding pathways can provide insights into ligand binding/unbinding kinetics and reveal potential protein “gateways” into active sites, which may be targeted for the binding of small molecules to block the active site. The Slab Mode of SILCS-MC docking allows users to dock ligands along a user-defined pathway.
In the Slab Mode of SILCS-MC docking, the user defines the “start” and “end” point (large green and red circles in the figure below, respectively) of a linear pathway for the ligand to be docked along. With the user-defined pathway as the axis, a series of cylindrical sampling regions (red cylinders in the figure below) are constructed with a tunable “radius” and “thickness” spanning the entire length between the start and end point. Users may optionally add additional sampling cylinders beyond the start (“reverse”) and end (“forward”) points (small green and red circles in the figure below, respectively). After running SILCS-MC with Slab Mode, the docked poses of the ligand along the pathway, in sequential order, will be generated (right panel of the figure below).
SILCS-MC Slab Mode Docking Using the CLI
Ligands can be docked along a user-defined pathway using the SILCS-MC Slab Mode docking through the following steps:
Launch SILCS-MC Slab Mode Docking:
To set up and run SILCS-MC docking using the Slab Mode from the command line interface, create a directory containing all the ligands to be evaluated. Each ligand can be stored as a separate SDF or Mol2 file. Alternatively, all the ligands can be combined into a single SD file. Additionally, determine the start and end point, in Cartesian coordinates, of the pathway along which the ligand will be docked. With this information, enter the following command to set up and launch the Slab Mode SILCS-MC docking run:
$SILCSBIODIR/silcs-mc/1_run_silcsmc_slab prot=<prot pdb> ligdir=<ligand directory> \ start=<"x,y,z"> end=<"x,y,z"> *Required parameters:*Path and name of protein PDB file:
prot=<protein pdb file>
Path and name of directory containing ligand SDF or Mol2 files:
ligdir=<ligand directory>
If the ligdir option is used, only one molecule per file under
ligdirwill be processed for SILCS-MC docking. Subdirectories underligdirwill also be processed. For an SD/SDF file containing multiple molecules, usesdfile=<path to sdfile>instead of theligdiroption.Starting point of the pathway along which the ligand(s) will be docked:
start=<"x,y,z"; e.g., start="20,30,-2">
End point of the pathway along which the ligand(s) will be docked:
end=<"x,y,z"; e.g., end="20,30,22">
Optional parameters:
Path and name of directory containing FragMaps:
mapsdir=<location and name of directory containing FragMaps; default=maps>
Name of the directory containing the SILCS-MC output:
silcsmcdir=<name of output directory; default=3_silcsmc>
Increments in which the ligand will be docked along the pathway (thickness of the slab):
thickness=<thickness/height of the slab; default=2.0 A>
Sampling radius for each docking run along the pathway (radius of the slab):
radius=<radius of the slab; default=5.0 A>
Extension of sampling beyond end point:
forward=<extra slab number of forward sampling after end; default=0>
Extension of sampling beyond starting point:
reverse=<extra slab number of reverse sampling before start; default=0>
Path and name of ligand SD file:
sdfile=<location and name of SD file, this option will overwrite ligdir>
If the
sdfileoption is used, theligdiroption is not needed and any input for theligdiroption will be overwritten. Thesdfileoption is recommended if the user has an SD file containing all ligands under investigation.
Evaluate docked poses:
The SILCS-MC tool set allows users to easily evaluate docked poses generated from the SILCS-MC Slab Mode docking runs. Users can collect the docked poses along the specified pathway using
2_calc_lgfe_min_avg_sdwith theslab=trueparameter (see Best-Pose Retrieval). In addition, using2_calc_lgfe_min_avg_sdwill additionally collect and rank order the resulting docked poses. SILCS simulation snapshots in which the protein conformation best complements the refined conformation and orientation of the ligand using can also be extracted with3_scan_traj(see Best Protein–Ligand Complex Retrieval).For details on evaluating the resulting SILCS-MC Slab Mode docked poses, please refer to Assessment of SILCS-MC Docked/Refined Poses in CLI.
SILCS-MC Slab Mode Docking Protocol Details
SILCS-MC Slab Mode docking spawns a single gpu job per ligand. In SILCS-MC Slab Mode
docking, the pathway along which the ligand(s) will be docked is divided into cylindrical
slabs. By default, the cylindrical slabs are 2 Å thick with a radius of 5 Å. The
thickness and radius of the cylindrical slabs can be customized in the command line
interface (thickness and radius optional parameters; see above). For each
cylindrical slab, a maximum of 250 cycles and a minimum
of 50 cycles of Monte Carlo/Simulated Annealing (MC/SA) sampling of the ligand
within the cylindrical slab is performed. Each of these 250 cycles consists of
10,000 steps of MC at a high temperature followed by 40,000 steps of SA towards a lower
temperature. At the begining of each cycle, the ligand will be reoriented within the
cylindrical slab. The MC sampling has three types of moves: i) molecular translations
with a maximum step size of 1.0 Å, ii) molecular rotations with a maximum step
size of 180.0°, and iii) intramolecular dihedral rotations with a maximum step size
of 180.0°. For intramolecular dihedral rotations, only the rotatable dihedral angles
are selected for MC moves. The lowest LGFE scoring pose from the MC sampling is used
as a starting pose in the following SA sampling. The SA sampling also involves the
same three types of moves, but with a smaller step size compared to the MC sampling:
i) molecular translations with a maximum step size of 0.2 Å, ii) molecular rotations
with a maximum step size of 9° and, iii) intramolecular dihedral rotations with a
maximum step size of 9°.
For each cylindrical slab, after a minimum of 50 MC/SA cycles, if the LGFE score
difference between the top three poses (defined by lowest LGFE scores) are less than 0.5
kcal/mol, then the run is considered converged and terminated. If the top three scored
poses are separated by more 0.5 than kcal/mol, then the MC/SA procedure continues
either until the convergence criterion is met or until a maximum of 250 MC/SA cycles
have been completed. At the end of all cycles per cylindrical slab, the lowest LGFE
scoring pose for each cylindrical slab is saved and stored in a multi-frame SD file
(3_silcsmc/1/out/<lig>.sdf) with each frame corresponding to the lowest LGFE scoring
pose of a given cylindrical slab along the user-defined pathway.
Assessment of SILCS-MC Docked/Refined Poses in CLI
Upon completion of the SILCS-MC docking and/or refinement simulations, the resulting docked poses can be assessed using tools available with your SilcsBio software. To extract the structures of docked poses and evaluate their LGFE scores (energetic favorability) and 4DBA scores (bioavailability), follow the instructions in Best-Pose Retrieval. To extract protein conformations that complement SILCS-MC docked structures and generate a protein–ligand complex structure, follow the instructions in Best Protein–Ligand Complex Retrieval after extracting the docked poses through Best-Pose Retrieval.
Best-Pose Retrieval
Once the SILCS-MC simulation is finished, users can retrieve the LGFE scores for each ligand subjected to SILCS-MC using the following command:
$SILCSBIODIR/silcs-mc/2_calc_lgfe_min_avg_sd ligdir=<directory containing ligand mol2/sdf>
The above command will rank order and collect the most energetically favorable (lowest
LGFE) docked poses resulting from the SILCS-MC docking runs. By default, the corresponding
LGFE scores per ligand will be output in the <silcsmcdir>/lgfe.csv file (see
below). The most energetically favorable binding pose(s) will be
stored in the <silcsmcdir>/minconfpdb/ folder in SDF file format.
Below is an example of the contents of the lgfe.csv output from this script:
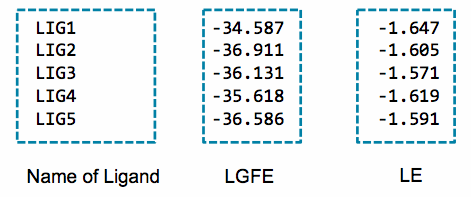
An alternative to the LGFE score is the ligand efficiency (LE). The LE is calculated as the LGFE score divided by the number of heavy atoms in each ligand.
Required parameter:
Path and name of directory containing ligand Mol2/SDF files:
ligdir=<location and name of directory containing ligand mol2/sdf>
The same directory containing the ligand Mol2/SDF files specified with
ligdirin the previous1_run_silcsmc_*step should be used if the poses for all docked ligands are being collected. If only the poses for a subset of the docked ligands is desired, then a new directory containing the Mol2/SDF files of the subset of ligands can be specified. Ifsdfilewas used in the previous step, use the samesdfileparameter for this step.
Optional parameters:
Name of the directory containing the SILCS-MC output:
silcsmcdir=<name of output directory; default=3_silcsmc>
The
silcsmcdirshould correspond to thesilcsmcdirspecified in the previous1_run_silcsmc_exhaustivestep. If nosilcsmcdirwas specified in the previous step, then the defaultsilcsmcdir=3_silcsmcshould be used.Path and name of ligand SD file:
sdfile=<location and name of SD file, this option will overwrite ligdir>
The same SD file should be used as in the previous
1_run_silcsmc_exhaustivestep. If thesdfileoption is used, theligdiroption is not needed and any input for theligdiroption will be overwritten. Thesdfileoption is recommended if the user has an SD file containing all ligands under investigation.Number of output poses:
npose=<number of best scoring poses to collect; default=1>
By default, only the best scoring, based on LGFE, docked pose will be output. To ouput additional docked poses, use the
nposeparameter and specify the desired number of docked poses. E.g., if 5 poses are desired, usingnpose=5will result in the 5 best scoring poses being output.Collect diverse poses based on RMSD clustering:
rmsd_cluster=<when npose more than 1, cluster by RMSD; true/false; default=false>
When
rmsd_cluster=trueandnposeis greater than 1, the next best scoring poses will be selected if it is not within the RMSD cutoff of the any of the previously selected poses. The RMSD cutoff is set to 3.0 Å by default and <rmsd_cluster_id> is already calculated during the SILCS-MC docking. Here we are just that information to select diverse poses.Option to output docked poses in PDB format:
pdb=<write PDB file in minconfpdb folder; true/false; default=false>
When
pdb=truethe docked poses will be output in PDB format in addition to the default SDF file format.Option to output ligand SMILES strings:
smiles=<write SMILES string to lgfe.csv; true/false; default=false>
When
smiles=truethe outputlgfe.csvfile will include the ligand SMILES strings in the second column of the file.Option to output RMSD w.r.t input pose:
ligrmsd=<write ligand RMSD w.r.t. input pose to lgfe.csv; true/false; default=false>
When
ligrmsd=truethe outputlgfe.csvfile will include a LigRMSD column and LigCOMD column. The LigRMSD column will be populated with the RMSD of the docked pose with respect to the input pose. The LigCOMD will be populated with the distance between the center of geometry of the docked pose and the input pose. The input pose is the ligand conformation and orientation in the input Mol2/SDF file. Note that this RMSD is calculated during the SILCS-MC docking run and is simply printed into the output file with no additional calculations performed; providing a different input pose in this step will not change the LigRMSD or LigCOMD values.Option to calculate 4DBA descriptors and 4DBA-LGFE scores:
fourdba=<true|false; default=false>
When
fourdba=truethe four 4DBA descriptors of each input ligand will be calculated and output intolgfe-4dba.csv. A high 4DBA score indicates that the ligand is bioavailable while a low 4DBA score indicates that the ligand is not bioavailable. For more information of 4DBA and LGFE-4DBA scores, please see 4D Bioavailability (4DBA) Calculation.Path to python executable:
python=<path to python executable for 4DBA calculations>
The default python path can be checked using the
which pythoncommand. Therdkit,tqdm,ipythonandscipypackages must be installed for the 4DBA score calculation. Please refer to Python 3 Requirement for more information.Option to extract LGFEs and poses along a pathway for Slab Mode (when
1_run_silcsmc_slabwas used to dock):slab=<true|false; default=false>
When
slab=true, an additional output filelgfe_slab.csvwill be produced. This file will contain the ligand name, step along the pathway (slab), and the ligand LGFE, ligand efficiency (LE), and center-of-geometry at that step. In conjunction withsmiles=true, the ligand SMILES strings will also be output into the second column of thelgfe_slab.csvfile. In addition, an SD file containing the docked poses of the ligand along the user-defined pathway will be stored for each ligand in3_silcsmc/conf_slab/.Option to sort the output by LGFE:
sortbylgfe=<sort lgfe.csv output by LGFE; true/false; default=false>
When
sortbylgfe=true, the outputlgfe.csvfile will be sorted by LGFE in ascending order. By default, the outputlgfe.csvfile is sorted by the ligand name in alphanumeric order when ligdir is used and by the ligand index in the SD file when sdfile is used.Submit job to queuing system:
batch=<submit job to queuing system; true/false; default=false>
Secondary Ligand Parameters:
If secondary ligands were used in the SILCS-MC docking, a selection criteria can be
applied to the secondary ligands to determine which ligands to include in the output
lgfe.csv file. The default selection criteria was defined for water molecules
as secondary ligands and is adapted from ref. [18], in which SILCS-MC
docking with waters as secondary ligands was performed and validated against
crystal structures of protein–ligand complexes containing water mediated interactions.
Distance cutoff (in Å) for secondary ligand selection:
distcutoff=<cutoff for max distance to select seclig in Angstroms; default=4.0>
The
distcutoffparameter is the maximum distance allowed between the ligand and secondary ligand. By default, thedistcutoffis set to 4.0 Å.LGFE cutoff for secondary ligand selection:
lgfecutoff=<cutoff for max allowed LGFE of seclig (SLigLGFE); default=0.5>
The
lgfecutoffparameter is the maximum LGFE allowed for the secondary ligand. By default, thelgfecutoffis set to +0.5 kcal/mol.Total energy cutoff for secondary ligand selection:
totecutoff=<cutoff for max allowed TotE (IntE+SLigLGFE) of seclig; default=-0.6>
The
totecutoffparameter is the maximum total energy allowed for the secondary ligand. By default, thetotecutoffis set to -0.6 kcal/mol.Interaction energy cutoff for secondary ligand selection:
intecutoff=<cutoff for max allowed interaction energy (IntE) between lig-seclig; default=-1.1>
The
intecutoffparameter is the maximum interaction energy allowed between the ligand and secondary ligand. By default, theintecutoffis set to -1.1 kcal/mol. Note that the interaction energy is the sum of the electrostatic and Lennard-Jones interaction energies between the ligand and the secondary ligand to be selected.Sort the output by total LGFE:
sortbytotlgfe=<sort lgfe.csv output by TotLGFE; true/false; default=false>
When
sortbytotlgfe=true, the outputlgfe.csvfile will be sorted by the total LGFE in ascending order. By default, the outputlgfe.csvfile is sorted by the ligand name in alphanumeric order.
Best Protein–Ligand Complex Retrieval
After retrieving the best ligand pose from SILCS-MC, users can visualize it in the context of SILCS FragMaps. However, since FragMaps are typically visualized with the initial protein structure, the docked ligand pose may overlap with the protein structure. The best protein–ligand complex retrival tool allows users to retrieve simulation snapshots, in which the conformation of the protein (or RNA) best complements the docked ligand, from the SILCS simulations. The most complementary conformations of the protein or RNA are determined by the favorability of non-bonded interactions between the protein/RNA and the docked ligand.
To retrieve complementary conformations of the target protein/RNA, first extract the
protein/RNA trajectory from SILCS simulations by re-running the
silcs/2b_gen_maps step with the option traj=true:
${SILCSBIODIR}/silcs/2b_gen_maps prot=<prot pdb> traj=true
Additionally, use oldversion=true if v2023.1 or older versions was used to
to run 2a_run_gcmd step, i.e., the simulation trajectories were saved without
hydrogen atoms.
The above command will generate traj.xtc and traj.pdb files as output, which
will be used as input for the extraction of the best protein–ligand complex
structure(s).
The best protein–ligand complex retrieval tool is run using the below command.
The resulting output files will be top<n>_<prot>_<lig>.csv and
top<n>_<prot>_<lig>.pdb, where <n> is the user-defined top number of frames
to extract (defined with the top parameter), <prot> is the protein PDB name,
and <lig> is the ligand name. top<n>_<prot>_<lig>.csv will contain the
frame number from which the protein conformation originated from and, for each frame,
the electrostatic interaction energy, LJ interaction energy, and the total interaction
energy between the protein and the ligand at the corresponding frame.
top<n>_<prot>_<lig>.pdb will contain the ensemble of protein–ligand complex
structures.
${SILCSBIODIR}/silcs-mc/3_scan_traj prot=<prot pdb> ligdir=<ligand SDF/Mol2 directory>
Required parameters:
Path and name of protein PDB file:
prot=<prot pdb>
Path and name of directory containing ligand Mol2/SDF files:
ligdir=<ligand SDF/Mol2 directory>
Typically this
ligdirshould be pointing to theminconfpdbdirectory output from the previous2_calc_lgfe_min_avg_sdstep withnpose=1option, meaning that the SDF files in the ligdir are the best docked/refined poses of the ligands with one pose per sdf file.Alternatively, the
sdfileoption can be used to specify the location and name of the SD file containing all ligands under investigation.
Optional parameters:
Name of the output directory:
outputdir=<name of output directory; default=scan_traj>
Number of top frames to extract:
top=<number of top frames to extract; default=10>
The simulation snapshots are ranked by the interaction energy of the docked pose with the protein conformation extracted from the simulation snapshot. By default only the Lennard-Jones component of the interaction energy is considered, this can be changed using the
sortbyparameter. The lowest interaction energy conformation is considered the most energetically favored conformation and the corresponding frame will be ranked first.Number of frames to skip while processing trajectory:
skip=<number of frames to skip; default=100>
Interaction energy component(s) used to rank the frames:
sortby=<type of energy to sort frames by; ELEC/LJ/TOTAL; default=LJ>
Users can select from electrostatic (
ELEC), Lennard-Jones (LJ) or total (TOTAL) interaction energies. By default, the simulation snapshots are ranked based onLJ.Option to ignore hydrogen atoms:
noh=<ignore protein hydrogens for energy calculation; true/false; default=false>
Users can choose to perform the energy calculation with protein hydrogen atoms ignored. Ligand hydrogen atoms will never be ignored. When
noh=true, this calculation will be performed in addition to the calculation with hydrogens.Path and name of SD file:
sdfile=<location and name of SD file>
Only one molecule in each Mol2/SD file under
ligdirwill be processed. If you have SD file with multiple molecules, use thesdfileoption instead of theligdiroption.Warning
The calculation will automatically loop over all ligands in
ligdirorsdfile. If there are many ligands inligdirorsdfile, this calculation may require a long compute time, and ifoldversion=true, many output files may be produced.Number of threads to use:
nproc=<number of threads to use; default=4>
Specify calculation for v2023.x or older SilcsBio versions:
oldversion=<true|fase; default=false>
Use
oldversion=trueif the option was used to generate traj.xtc/pdb files insilcs/2b_gen_maps. This option should be set totrueif SilcsBio v2023.x or older versions were used to run the SILCS simulations (silcs/2a_run_gcmd).Tip
For users with access to both the SILCS-Small Molecule Suite and the CGenFF Suite, running standard MD simulation can be helpful to evaluate or refine the protein–ligand complex structure obtained from SILCS-MC Docking or Pose Refinement. The structure retrieved from the process described in Best Protein–Ligand Complex Retrieval may be used to create the initial protein–ligand complex structure for further refinement using MD simulations. For more information on running standard MD simulations through the CGenFF Suite, please refer to Standard Molecular Dynamics (MD) Simulations.
User-Defined Protocols through the CLI
In addition to the default docking, pose refinement, and pose evaluation protocols,
users can define their own SILCS-MC protocols. To do so, copy
${SILCSBIODIR}/templates/silcs-mc/params_custom.tmpl to the location where
you intend to run your custom SILCS-MC protocol. Edit this copy to reflect your
customization; see below for a detailed description of user-definable
parameters. Parameter values in angle brackets in this file, such as
<SILCSBIODIR>, will be replaced automatically at runtime where possible.
After you have edited params_custom.tmpl, use the following command to set
up and run SILCS-MC. Each input ligand can be stored as a separate SDF or Mol2
file. Alternatively, all input ligands can be combined in a single SD file.
${SILCSBIODIR}/silcs-mc/1_run_silcsmc_custom prot=<prot pdb> \
ligdir=<directory containing ligand sdf/mol2> \
mapsdir=<directory containing SILCS FragMaps> \
paramsfile=<params_custom.tmpl>
The number of runs that will be spawned can also modified with the command-line
parameter totruns:
${SILCSBIODIR}/silcs-mc/1_run_silcsmc_custom prot=<prot pdb> \
ligdir=<directory containing ligand sdf/mol2> \
mapsdir=<directory containing SILCS FragMaps> \
paramsfile=<params_custom.tmpl> \
totruns=<# of runs>
Note
.sdf, .sd, or .mol2 files can be placed in the ligdir
directory, and SILCS-MC will read a single molecule from each file.
If a file contains multiple molecules, use of the ligdir
option will result in only the first molecule in the file being processed.
If you have an SD file with multiple molecules in it, replace
ligdir=<directory containing ligand mol2/sdf> with
sdfile=<path to sdfile> to process all molecules in the file.
Warning
Ligands, regardless of file format, must include all hydrogens, including pH-appropriate (de)protonations, and must have reasonable three dimensional conformations.
The full list of user-definable parameters for params_custom.tmpl is:
CGENFF_RULES <cgenff rules_file>(required)This file is needed by the internal CGenFF library to determine the correct force-field parameters for the ligand. The default value is
${SILCSBIODIR}/data/cgenff/cgenff.rulesCGENFF_PAR <cgenff parameter file>(required)Along with the CGENFF_RULES file, this file is needed by the internal CGenFF library to determine the correct force-field parameters for the ligand. The default value is
${SILCSBIODIR}/data/cgenff/par_all36_cgenff.prmSILCS_RULES <silcs rules file>(required)This rules file is the atom classification scheme (ACS) used to map the different atoms in the ligand to the corresponding SILCS FragMap types. This mapping is used to determine the appropriate “field” that will be applied to the different atoms in the ligand when attempting an MC-move. The default value is
${SILCSBIODIR}/data/silcs/silcs_classification_rules_2021_generic_apolar_scale_1f.datWhen
silcs_classification_rules_2021_generic_apolar_scale_1f.datis used, ligand atoms are assigned using generic classifications for mapping back to the FragMaps.Additional rules files are available in the
${SILCSBIODIR}/data/silcs/for other mapping schemes, including for more specific classifications and for using halogen FragMaps (see Setup with halogen probes).GFE_CAP <default: 3.0>Maximum allowable unfavorable GFE (kcal/mol) in the MC calculation.
RDIE <default: true>When true, the distance dependent dielectric (RDIE) scheme is used to treat intramolecular electrostatics. When false, CDIE (constant dielectric scheme) is used.
DIELEC_CONST <default: 4>Dielectric constant used in the intramolecular electrostatic interactions calculations.
MINIMIZE_INPUT <default: false>Perform minimization of input structure.
MINIMIZE_BFGS <default: false>Perform minimization of input structure using BFGS algorithm.
MIN_STEPS <default: 10000>Maximum number of steps of minimization performed using the steepest-descent algorithm with the ligand, before initiating MC simulation.
EMTOL <default: 0.01>Minimization is converged when the diff in total energy (totE) across the last 10 steps is smaller than this value. Once this criteria is satisfied minimization terminates.
MC_MOVE_RANGE <default:1.0 180.0 180.0>.Maximum range of translation, rigid body rotation and dihedral rotation per step of MC simulation.
MC_PRNT_FRQ <default: 0>Number of intermediate steps of MC to be written into OUTMCPDBFILE.
MC_STEPS <default: 10000>Number of steps of MC simulation to be performed per cycle.
MC_RUN <default: 1>Number of MC simulation cycles to be performed. By default, SILCS-MC performs one cycle of MC simulation with
MC_STEPS. In some cases, when users want to generate poses and orientations, and then perform a local refinement for each pose, they can setMC_RUNto a higher value.MC_STEPS2 <default: 10000>Number of steps of MC simulation to be performed for later cycle of
MC_RUN, only whenMC_RUNis larger than 1. By default, SILCS-MC performs one cycle (MC_RUN= 1) of MC simulation withMC_STEPS, andMC_STEPS2will be ignored.MC_MOVE_RANGE2 <default:0.1 90.0 60.0>.Maximum range of translation, rigid body rotation and dihedral rotation per step for
MC_STEPS2.SIM_ANNEAL_MOVE_RANGE <default:0.2 9.0 9.0>Maximum range of translation, rigid body rotation and dihedral rotation per step of simulated annealing simulation after MC simulation.
SIM_ANNEAL_STEPS <default: 40000>Number of steps of simulated annealing to be performed per cycle.
INIT_RUNS <default: 50>Number of MC/SA cycles before initiating checks for convergence.
NUM_TOL <default: 3>Number of top-scoring cycles with differences in LGFE less than DELTAE_TOLERANCE, before this simulation (run) is considered converged.
DELTAE_TOLERANCE <default: 0.5>When differences in LGFE of NUM_TOL most-favorable cycles are less than this defined tolerance value, convergence is reached and the program exits
DELTAE_BUFF <default: 10>Progression of MC+SA from one cycle to next is such that LGFE (of lowest conf) from MC should be less than (prev_min+deltae_buff). This ensures that N cycles are proceeding towards a minimum lower than that previously discovered lowest energy conformation.
TOTE_CRITERIA <default: false>When true, instead of LGFE, total energy (totE) of the system is used for convergence checks. Useful when running vacuum-phase MC simulations of the ligand.
TOT_RUNS <default: 250>Maximum number of MC simulation cycles. The program terminates if the
DELTAE_TOLERANCEcriteria is satisfied before reachingTOT_RUNS. Alternately, even if theDELTAE_TOLERANCEcriteria is not satisfied when the number of cycles executed reachesTOT_RUNS, the program terminates. The GPU version of SILCS-MC ignoresDELTAE_TOLERANCEand runs for allTOT_RUNScycles specified.RANDOM_SEED: <default: system-time>Seed used in MC simulation. When not set, system-time is used as a seed.
SIMULATION_CENTER: <x,y,z>Cartesian coordinates of where the MC simulation should be performed.
SIMULATION_RADIUS: <default: 10.0 A>Radius of the sphere within which MC simulation will be performed.
RANDOM_INIT_ORIENT: <true/false>When set to
TRUE,SIMULATION_CENTERshould also be set. The ligand is placed within a sphere of sizeSIMULATION_RADIUSin a random orientation and a random conformation.When set to
FALSE, the center-of-geometry of the ligand is used as the center for the MC simulation, and the input pose of the ligand is used as the starting pose for every MC run. This is useful when the ligand pose in the pocket is well known.ATOM_TO_RESTRAIN: <atom number in sdf/mol2>When set, a spherical potential is applied to restrain the defined atom within the sphere during MC moves. This enables geometrically restraining a particular pharmacophore feature. Note, when using this feature, supply the full molecule with explicit hydrogens already added. Rrandom pocket pose and placement using
RANDOM_INIT_ORIENTtrue is incompatible with this option.When not set, the entire molecule is free to rotate/move/translate
ATOM_RESTRAINT_CENTER: <x,y,z>To be used in conjunction with
ATOM_TO_RESTRAINoption. This value is used to defined the center of the spherical potential.ATOM_RESTRAINT_RADIUS: <default: 1.0 A>To be used in conjunction with
ATOM_TO_RESTRAINoption. This value is used to defined the radius of the spherical potential. When not defined, then a default of 1 A is used.OUTRMSDFILE <output RMSD file>This file stores the RMSD and LGFE of the lowest energy conformation from each run of the MC/SA simulation. To be used in conjunction with
RANDOM_INIT_ORIENTset totrue.SILCSMAP <MapType> <map name> <scaling factor>(required)Multiple
SILCSMAPentries are typically defined, with each entry pointing to a FragMap filename<map name>.<scaling factor>is used to scale atomic Grid Free Energies (GFEs) for<MapType>atoms in the ligand being scored.<MapType>entries here must correspond to those defined in theSILCS_RULESrules file described above.OUTPUT_FORMAT [SDF|PDB](required)Choice of output format. SDF or PDB is supported.
OUTPUT_FILE <output file name>(required)This file stores the lowest energy conformation from each cycle of the MC/SA simulation.
LOGFILE <output log file>This file stores the energy statistics of the lowest energy conformation from each cycle of the MC/SA simulation.
When docking secondary ligands, use the parameters below:
MC_MOVE_RANGE_SECLIG <default: 1.0 180.0 180.0>Maximum range of translation, rigid body rotation and dihedral rotation per step of MC simulation for secondary ligands.
SIM_ANNEAL_MOVE_RANGE_SECLIG <default:0.2 9.0 9.0>Maximum range of translation, rigid body rotation and dihedral rotation per step of simulated annealing simulation after MC simulation for secondary ligands.
RANDOM_INIT_ORIENT_SECLIG <true/false>When set to
true, the secondary ligand is placed within a sphere in a random orientation. The radius and center of the sphere is defined bySIMULATION_RADIUS_SECLIGandSIMULATION_CENTER_SECLIG, respectively.When set to
false, the input pose of the secondary ligand is used as the starting pose for every MC run.SIMULATION_CENTER_SECLIG: <x,y,z>Cartesian coordinates of spherical sampling regions where the MC simulation should be performed for secondary ligands. This is used when
RANDOM_INIT_ORIENT_SECLIGis set totrue.SIMULATION_RADIUS_SECLIG: <default: 10.0 A>Radius of the sphere within which MC simulations will be performed for secondary ligands. This is used when
RANDOM_INIT_ORIENT_SECLIGis set totrue. Users are recommended to set this value to a different value toSIMULATION_RADIUSand to baseSIMULATION_RADIUS_SECLIGon the length of longest axis of the primary ligand to allow sampling of the secondary ligands around the primary ligand.