Plotting Probe Data¶
The SilcsBio software provides a utility to monitor probe concentrations and excess chemical potentials (\(\mu_\mathrm{ex}\)) as a function of GCMC-MD cycle. The following python script will generate these plots:
python $SILCSBIODIR/utils/python/plot_probe_data.py --prot <Protein PDB File>
optional arguments:
-h, --help show this help message and exit
--prot PROT protein PDB file (Required)
--numsys NUMSYS number of runs (Default=10)
--halogen HALOGEN halgen probes True/False (Default=False)
--begin BEGIN first gcmc-md cycle (Default=1)
--end END number of gcmc-md cycles (Default=100)
--vertical VERTICAL plot together vertically/horizontally (Default=False)
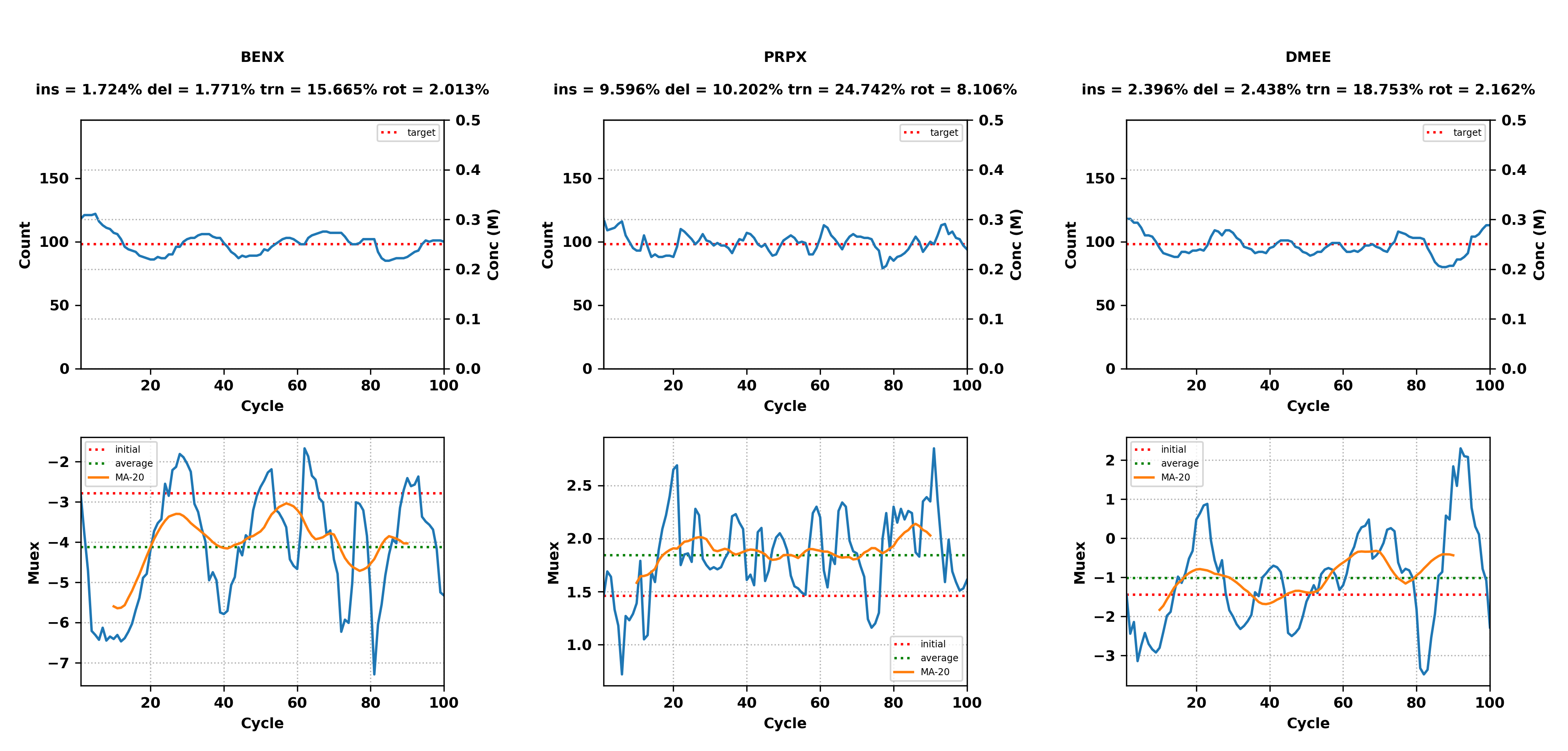
Below is an example of an output from running the python script:
1 BENX Acceptance: ins = 1.724% del = 1.771% trn = 15.665% rot = 2.013%
1 PRPX Acceptance: ins = 9.596% del = 10.202% trn = 24.742% rot = 8.106%
1 DMEE Acceptance: ins = 2.396% del = 2.438% trn = 18.753% rot = 2.162%
1 MEOH Acceptance: ins = 1.014% del = 1.037% trn = 13.012% rot = 0.680%
1 FORM Acceptance: ins = 0.772% del = 0.809% trn = 14.391% rot = 0.639%
1 IMIA Acceptance: ins = 1.183% del = 1.211% trn = 8.683% rot = 0.664%
1 ACEY Acceptance: ins = 1.453% del = 1.496% trn = 4.402% rot = 0.644%
1 MAMY Acceptance: ins = 3.865% del = 4.031% trn = 8.300% rot = 2.009%
1 SOL Acceptance: ins = 0.505% del = 0.506% trn = 10.704% rot = 1.026%
The output lists the SILCS simulation number, SILCS solute, and the accepted
insertions (ins), deletions (del), translations (trn), and
rotations (rot).
A detailed output for each simulation system will be stored in status.<system number>.txt
text file in the 2a_run_gcmd directory. An example of the contents of
status.<system number>.txt is shown below:
cycle fragname curr_N targ_N percent hfe muexdx vardx dxdx muex
1 benx 118 98 120.41 -0.790 1.000 0.000 0.000 -2.790
1 prpx 118 98 120.41 1.960 0.500 0.000 0.000 1.460
1 dmee 118 98 120.41 -1.440 1.000 0.000 0.000 -1.440
1 meoh 118 98 120.41 -5.360 2.000 0.000 0.000 -5.360
1 form 118 98 120.41 -10.920 2.500 0.000 0.000 -10.920
1 imia 118 98 120.41 -14.180 3.000 0.000 0.000 -14.180
1 acey 118 98 120.41 -97.310 10.000 0.000 0.000 -97.310
1 mamy 118 98 120.41 -68.490 7.000 0.000 0.000 -68.490
1 sol 21750 21750 100.00 -5.600 0.500 0.000 0.000 -5.600
2 benx 121 98 123.47 -0.790 1.000 -0.920 -0.300 -3.790
2 prpx 109 98 111.22 1.960 0.500 -0.220 0.450 1.690
2 dmee 118 98 120.41 -1.440 1.000 -1.600 -0.000 -2.440
...
99 imia 100 98 102.04 -14.180 3.000 -0.240 0.000 -7.160
99 acey 109 98 111.22 -97.310 10.000 0.000 1.000 -95.310
99 mamy 104 98 106.12 -68.490 7.000 0.000 0.700 -83.890
99 sol 21818 21750 100.31 -5.600 0.500 -0.031 0.000 -6.071
100 benx 100 98 102.04 -0.790 1.000 -0.080 0.000 -5.330
100 prpx 94 98 95.92 1.960 0.500 0.080 0.000 1.610
100 dmee 113 98 115.31 -1.440 1.000 -1.200 -0.000 -2.280
100 meoh 107 98 109.18 -5.360 2.000 -0.720 -0.400 -3.600
100 form 98 98 100.00 -10.920 2.500 2.500 0.000 -8.820
100 imia 109 98 111.22 -14.180 3.000 -1.320 -2.700 -11.180
100 acey 106 98 108.16 -97.310 10.000 -3.200 3.000 -95.510
100 mamy 105 98 107.14 -68.490 7.000 -1.960 -0.700 -86.550
100 sol 21793 21750 100.20 -5.600 0.500 -0.020 0.000 -6.090
In the status.<system number>.txt file, each row corresponds to a given solute
molecule in a given cycle. In each row, the following information is provided:
the cycle (
cycle),SILCS solute (
fragname),number of the solute molecule in the cycle (
curr_N),targeted number of the solute molecule (
targ_N),the percent number of the solute molecule over the targeted number of that solute molecule (
percent),the approximate hydration free energy in kcal/mol of the solute obtained from free energy perturbation simulations (
hfe) [10],the estimated magnitude of excess chemical potential variation (
muexdx),the standard variation in excess chemical potential (
vardx),the instantaneous variation in the excess chemical potential (
dxdx), andthe excess chemical potential (
muex).
More information on GCMC cycles and each variable is available in reference [27].
In addition, the python script will produce plots as .png files for each probe in
each simulation system. These plots will be stored in 2a_run_gcmd/acceptance_analysis/
as count.<protein PDB name>.<system number>.png. Below is an example of the output
plots: