Chemical Group Transformations¶
SILCS-MC ligand optimization and SSFEP both entail computations on derivatives of a parent ligand. Additionally, the user may wish to perform SILCS docking using a series of structures chemically related to a reference compound (parent ligand). Therefore, a task common to these use cases is that of performing chemical group transformations on a parent ligand to create a series of chemically-related compounds.
Ligand Modifications Using the SilcsBio GUI¶
The SilcsBio GUI provides an intuitive way to modify ligands directly in the context of SILCS-MC ligand optimization (see SILCS-MC Ligand Optimization Using the SilcsBio GUI) and SSFEP (see SSFEP Suite Quickstart). You can also use the SilcsBio GUI to perform chemical transformations independently of any other task by choosing Modify ligand for SILCS-MC from the Home page of the Small Molecule Suite. The utility is also available from the Home page of the SSFEP and CGenFF Suites through Modify ligand for SSFEP and Modify ligand for CGenFF
Output Mol2 files will be saved to a directory of your choosing on the computer where you are running the SilcsBio GUI. Instructions for building ligand modifications through Modify ligand for SILCS-MC/SSFEP/CGenFF from the Home page are provided below.
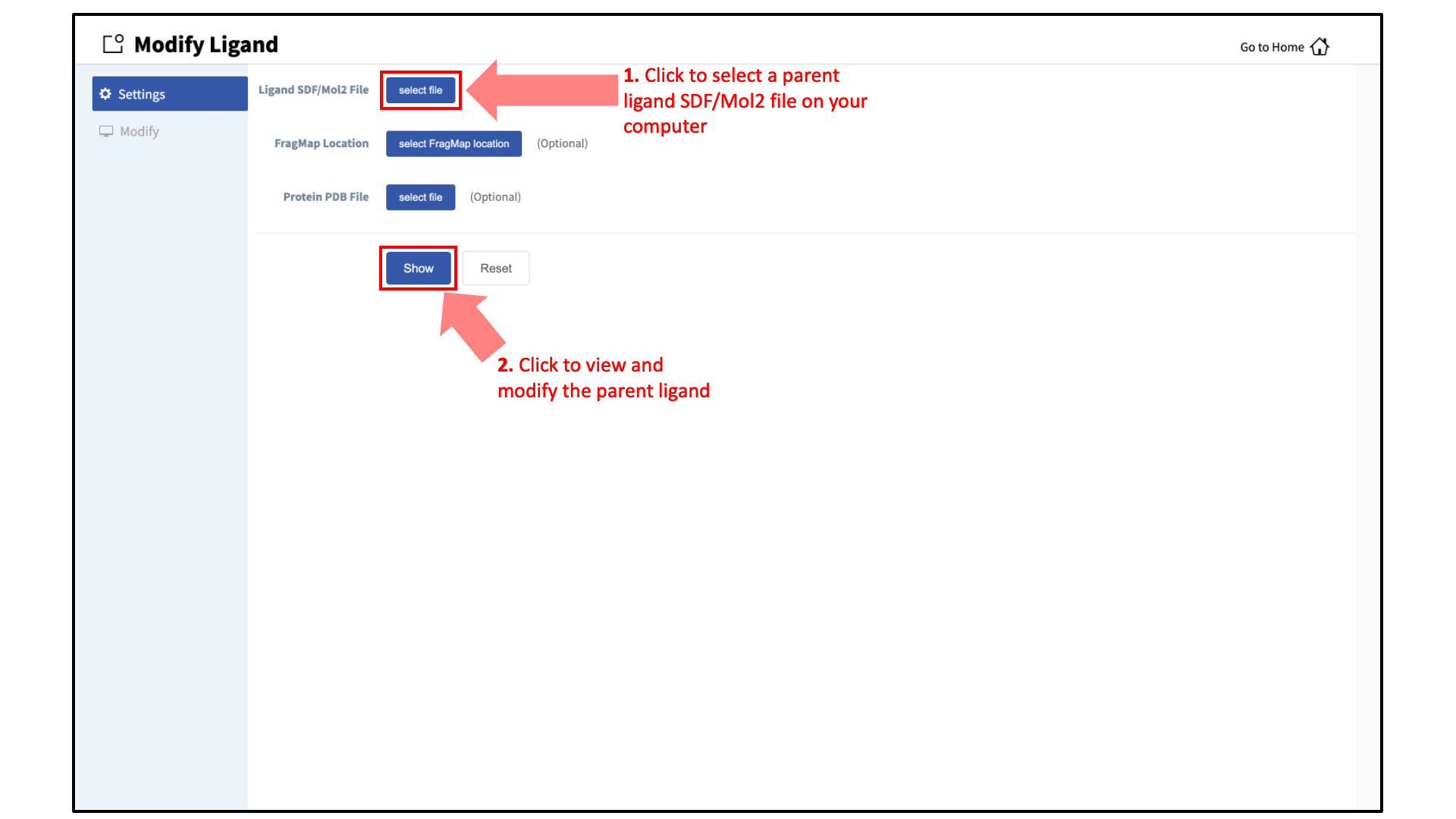
Enter the parent ligand SD/Mol2 file:
Provide the SD or Mol2 file of the parent ligand structure. To guide the selection of sites to modify and ligand modifications to apply, the user may also provide the struture of the target protein and the corresponding FragMaps. If the target protein and FragMaps are loaded, ensure that the parent ligand is appropriately positioned within the binding pocket. Click “Show” to proceed.
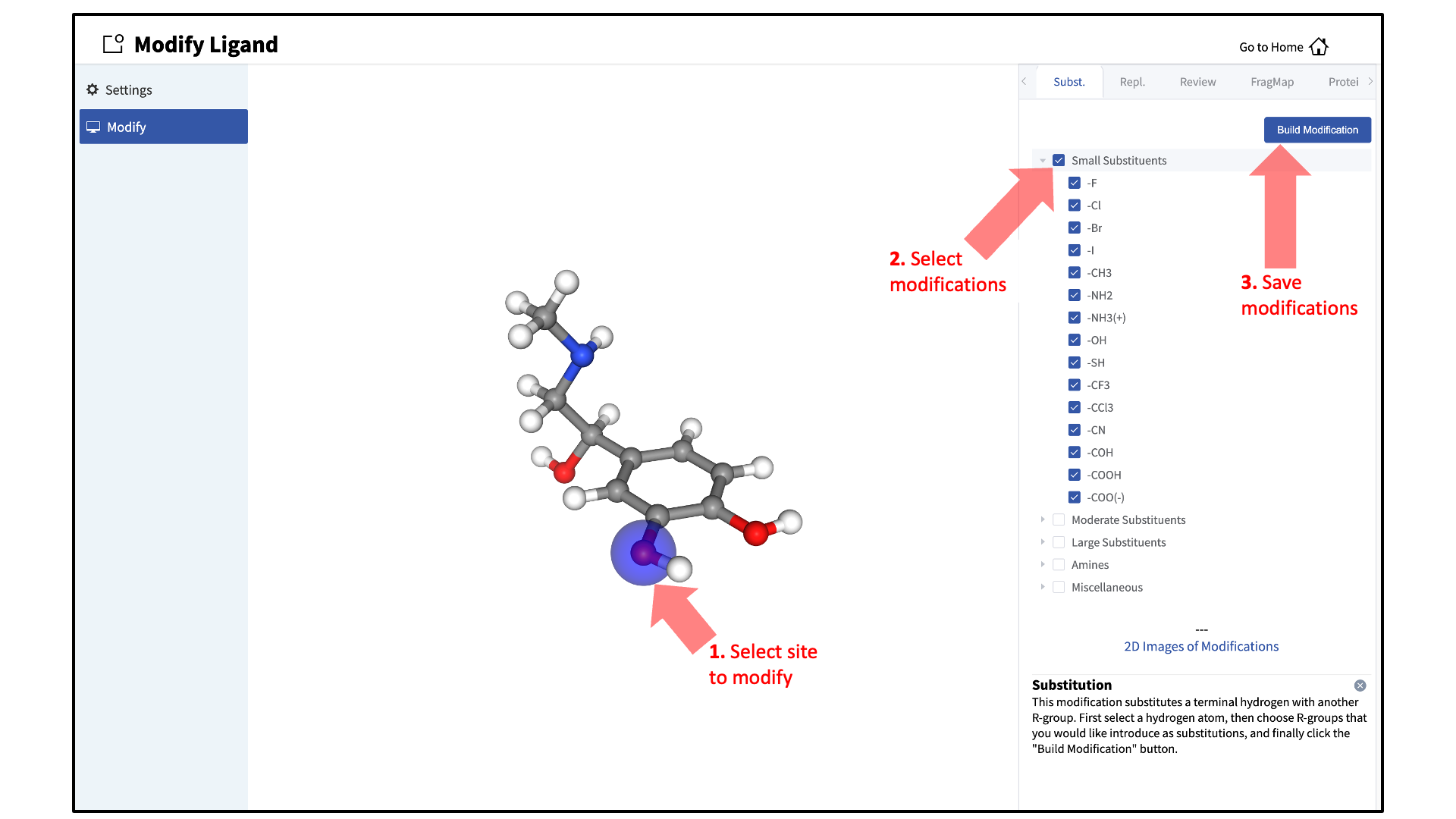
Add ligand modifications:
The GUI will display the parent ligand. First, select the atom to be modified. Then, select the desired modifications from the “Subst.” (Substitution) or “Repl.” (Replacement) tab in the right-hand panel. Substitution is used to substitute an atom with a functional group. Replacement is used to replace an atom in a ring or aliphatic acyclic group with another functional group that preserves the connectivity and valence of the ring or chain. The list of modification types in the GUI covers a very broad range of chemical functionality and size (see Available Ligand Modifications in the SilcsBio GUI for the list of possible modifications in SilcsBio GUI). Press the “Build Modification” button in the panel to build the selected modifications at the selected modification site.
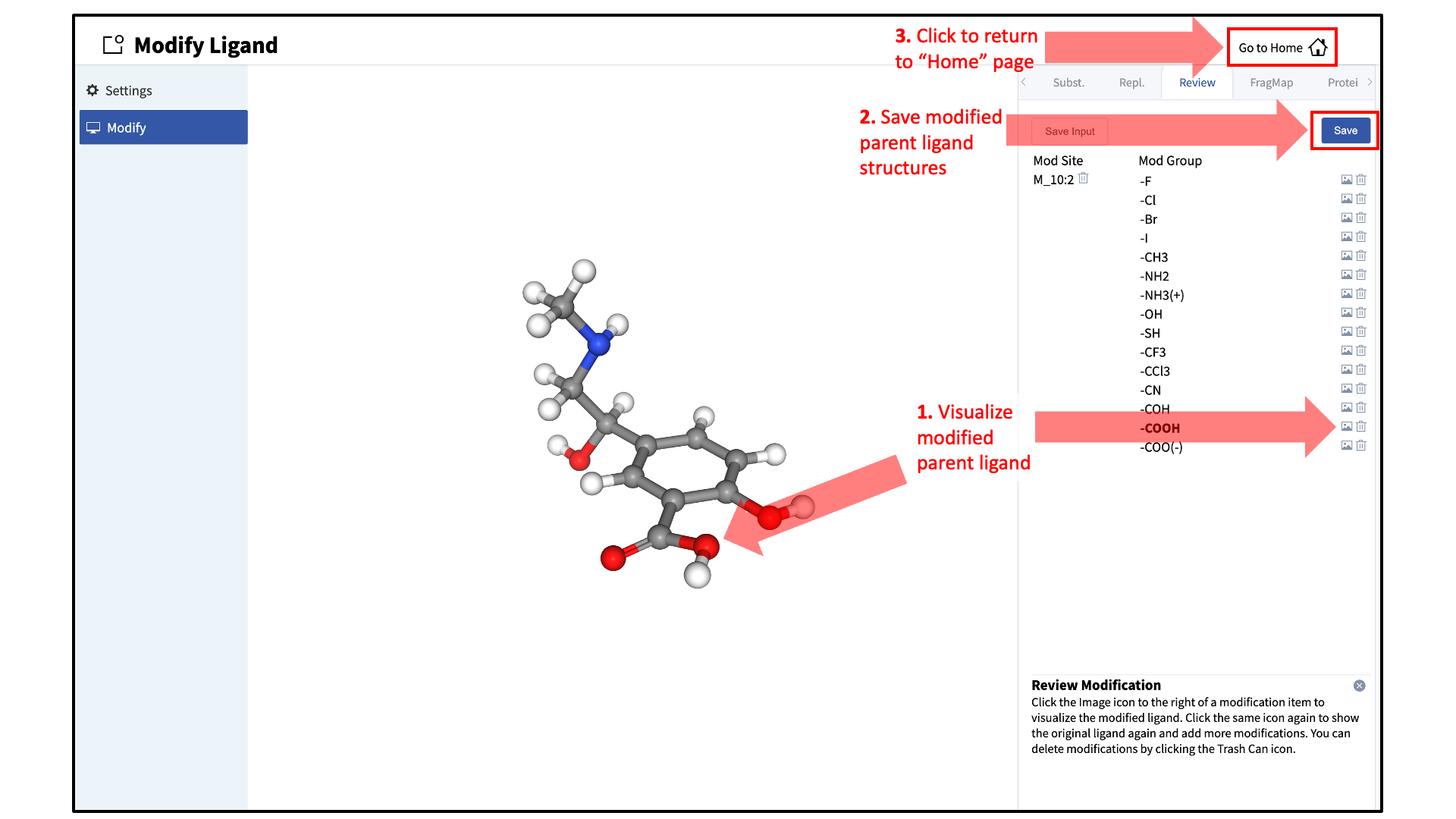
Save modified ligands:
Use the “Review” tab to confirm and save the desired modifications.
Valid modifications will have a small Image icon as well as a small Trash Can icon. Clicking on the Image icon will show the modification in the center panel. Clicking on it again will show the parent ligand. Clicking on the Trash Can icon will delete the proposed modification from the “Review” list. Additional modifications can be included in the list by re-entering the “Subst.” and “Repl.” tabs. Once the list of modifications is complete, save the modifications by clicking the “Save” button under the “Review” tab. Output Mol2 files will be saved to a user-specified directory of your choice on the local machine where the SilcsBio GUI is being run.
After saving the modified ligands, click “Go to Home” at the top right corner of the page to return to the “Home” page.
Available Ligand Modifications in the SilcsBio GUI¶
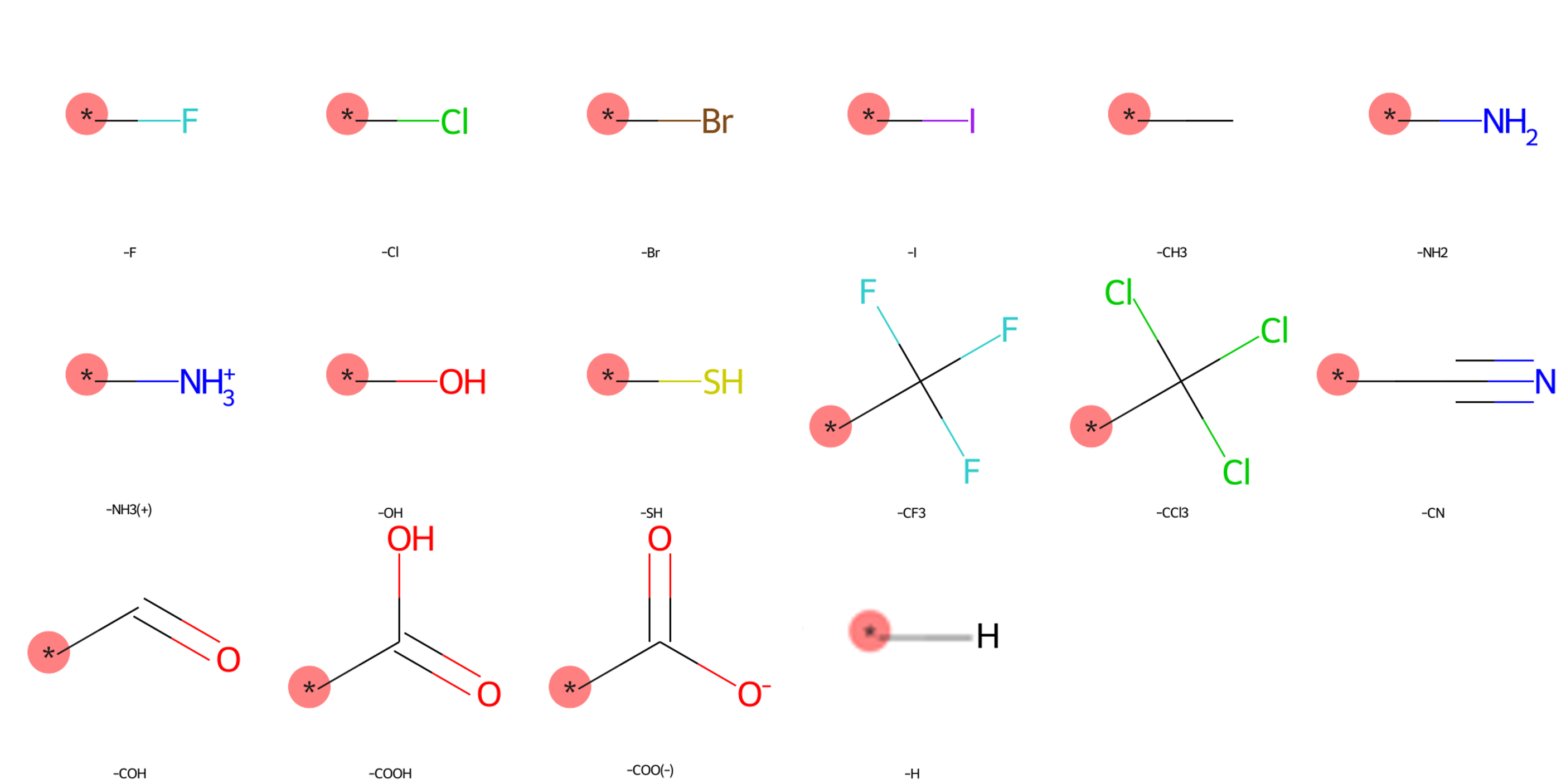
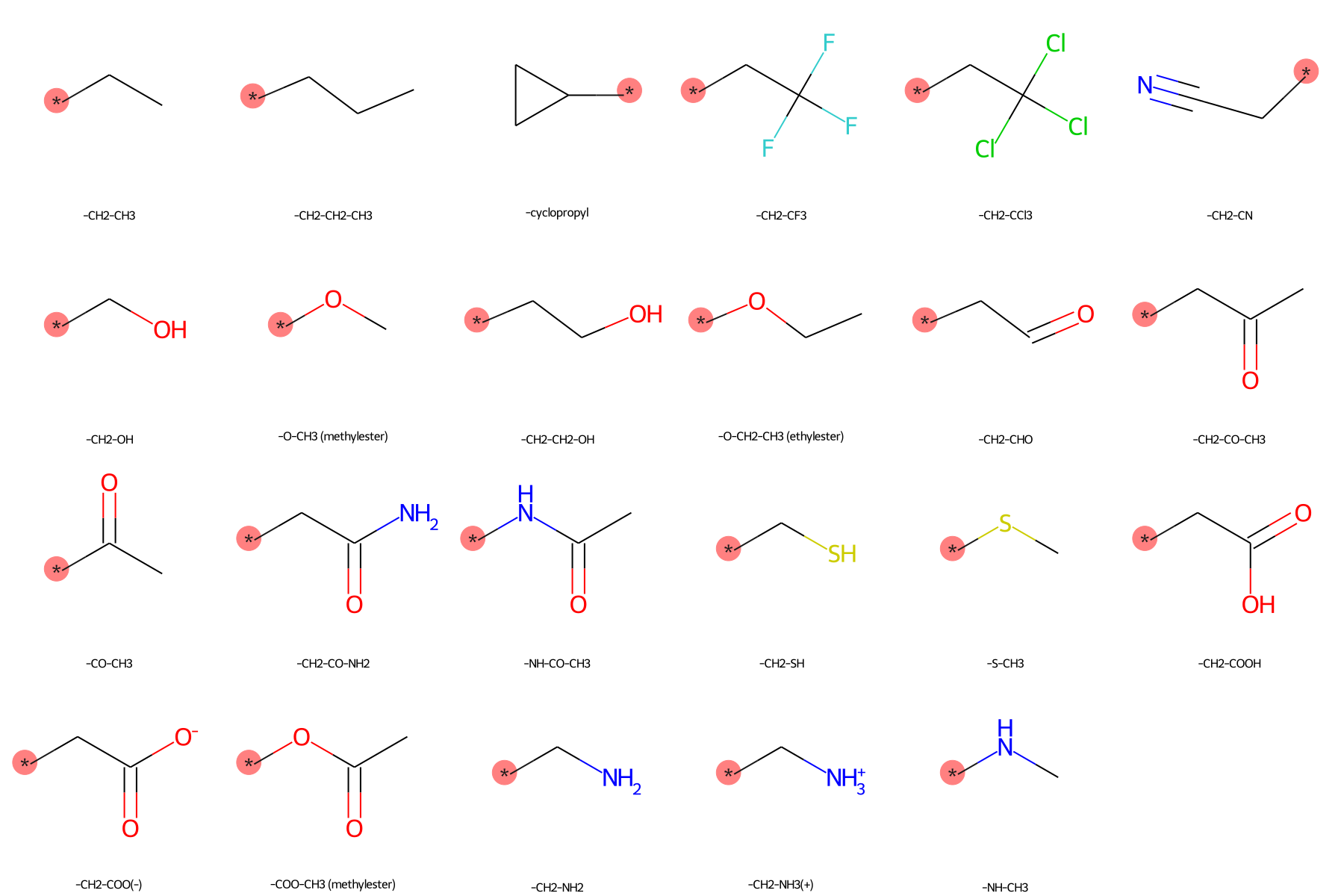
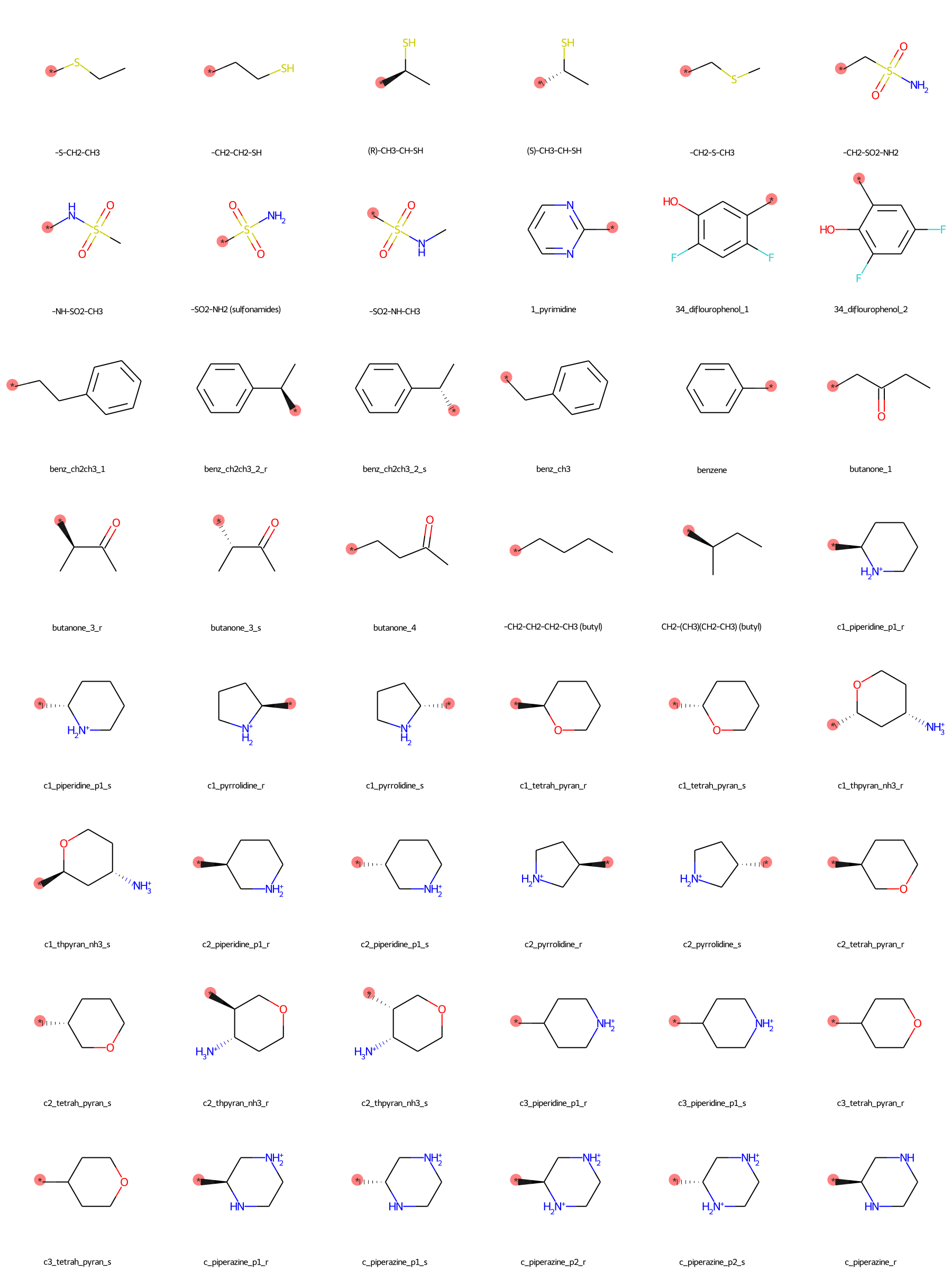
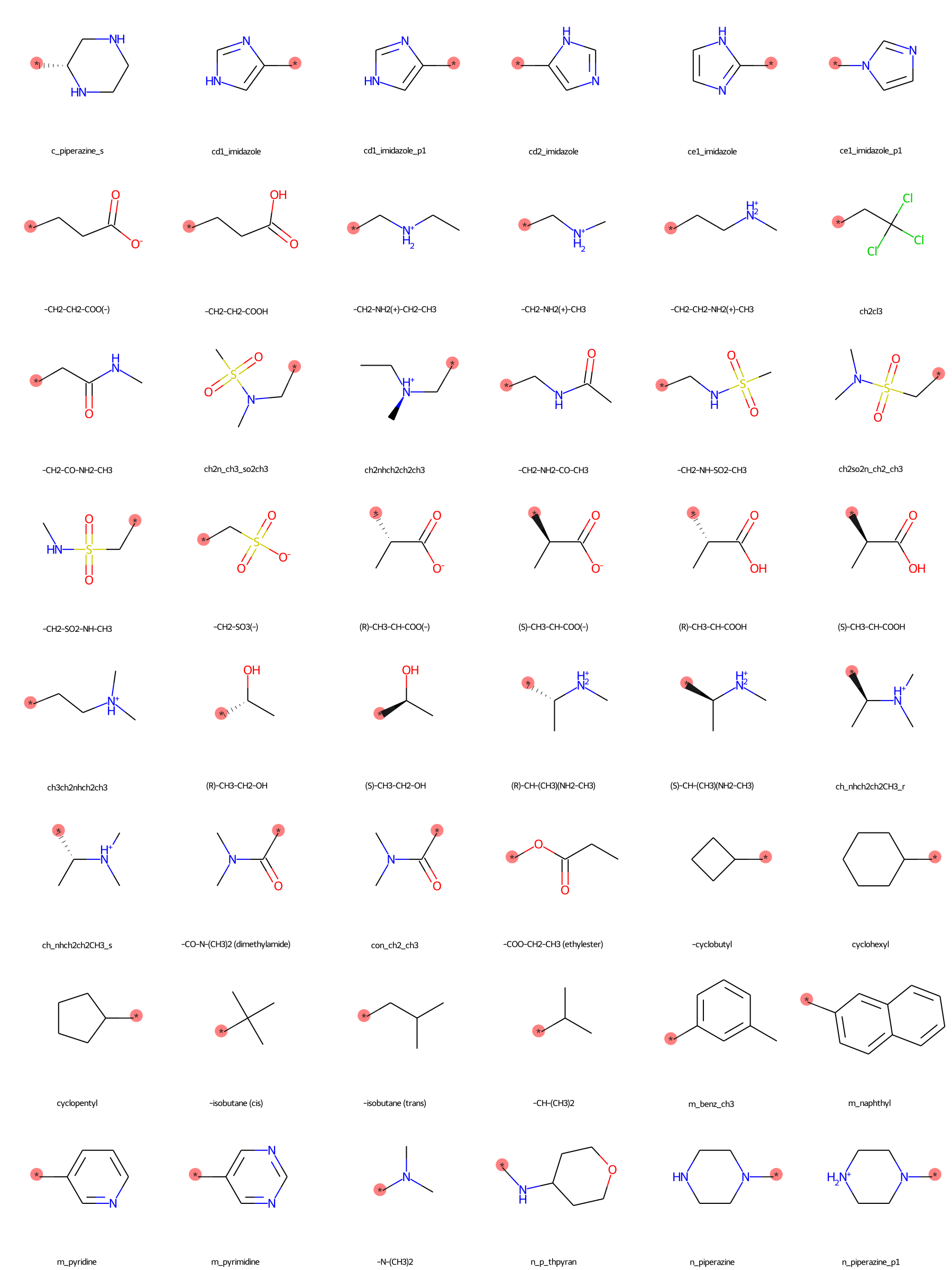
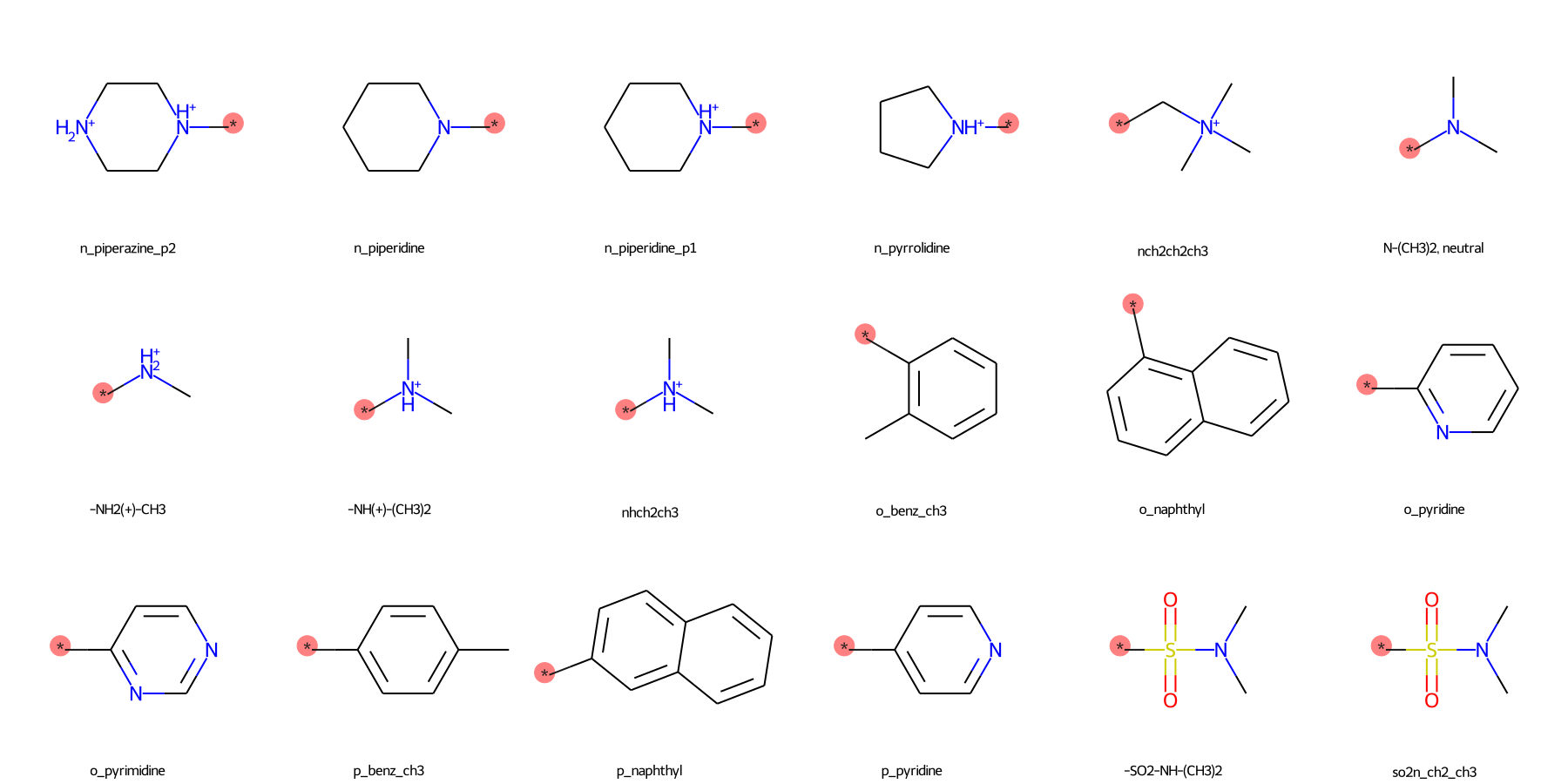
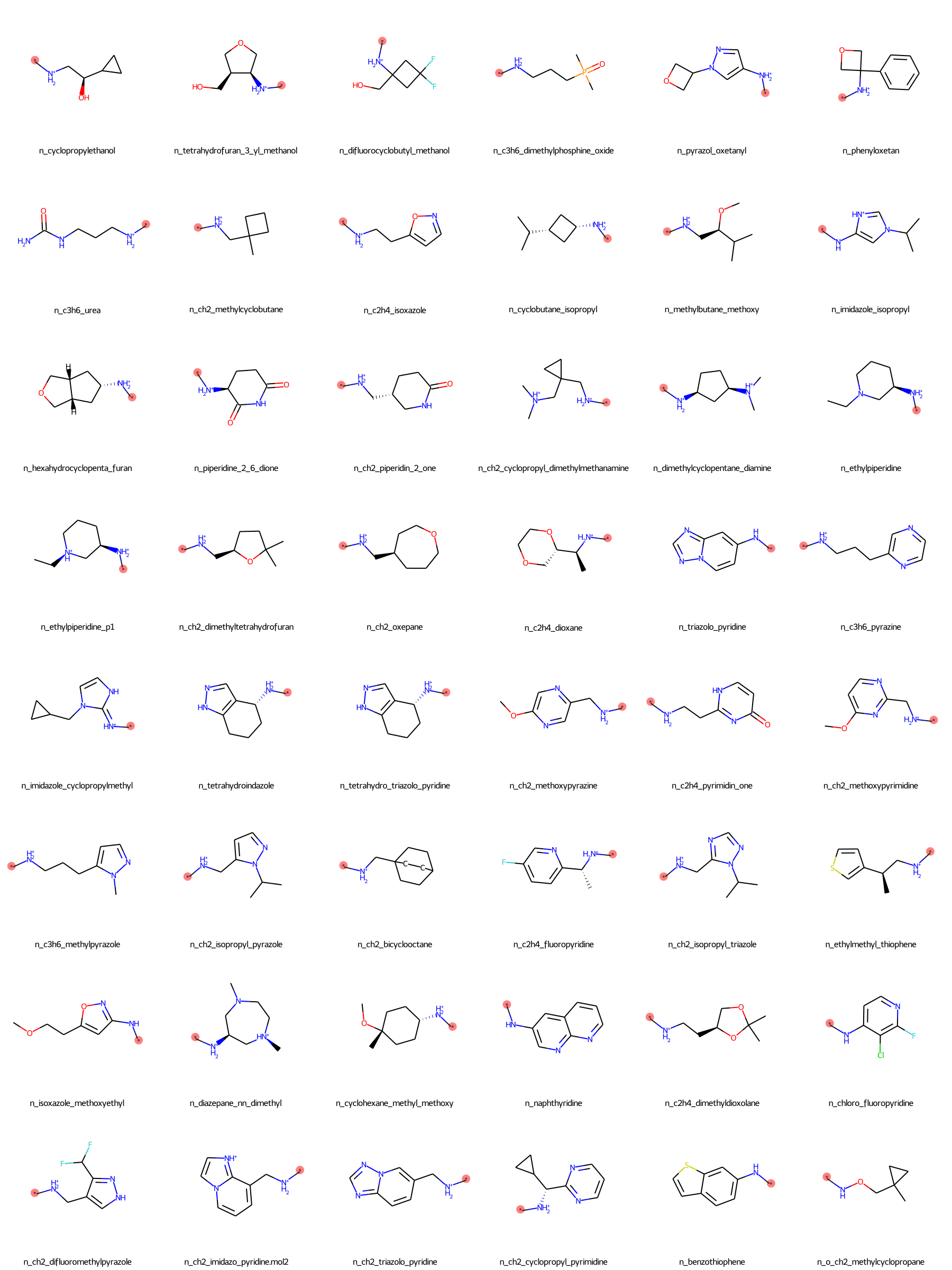
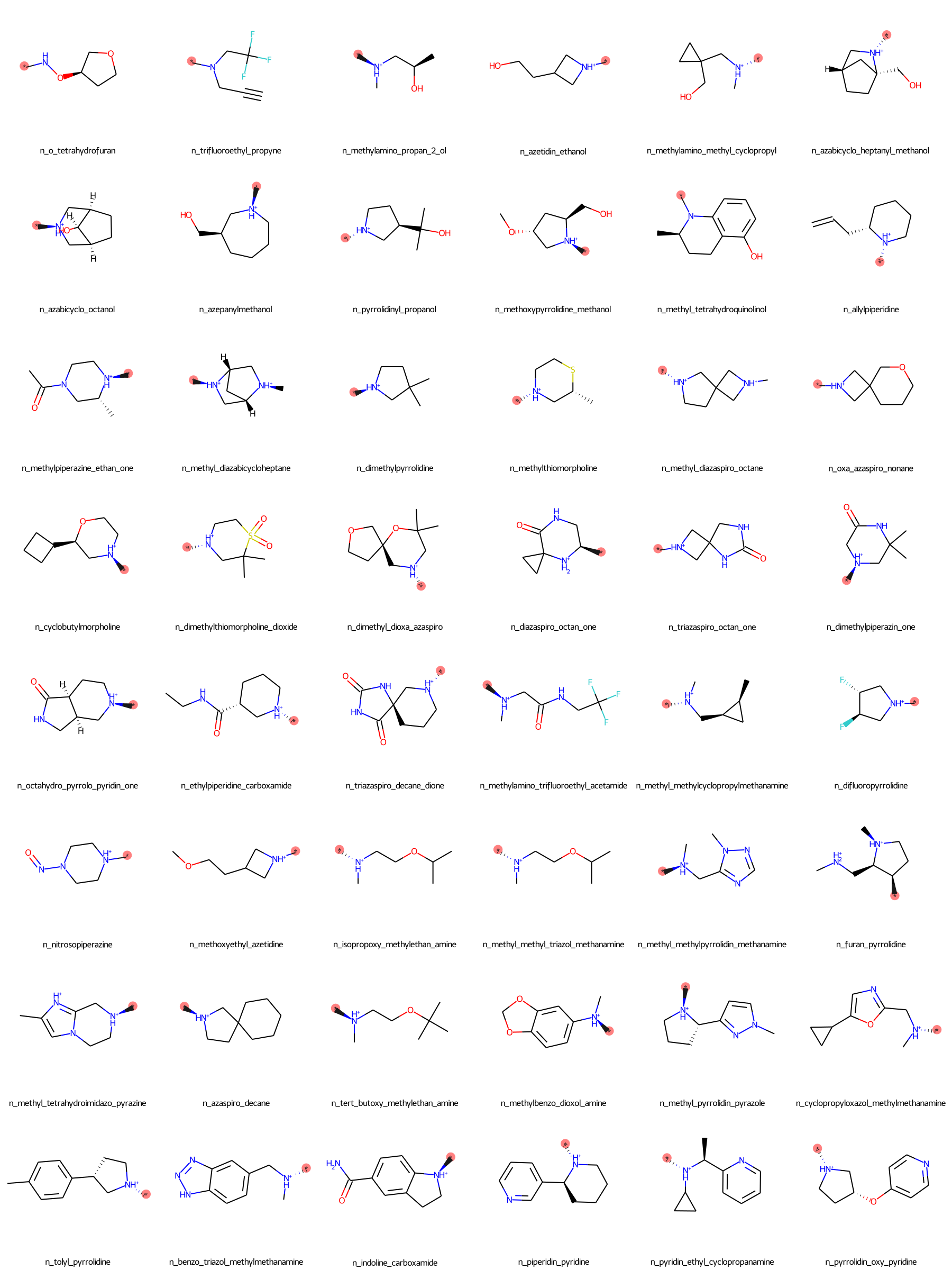
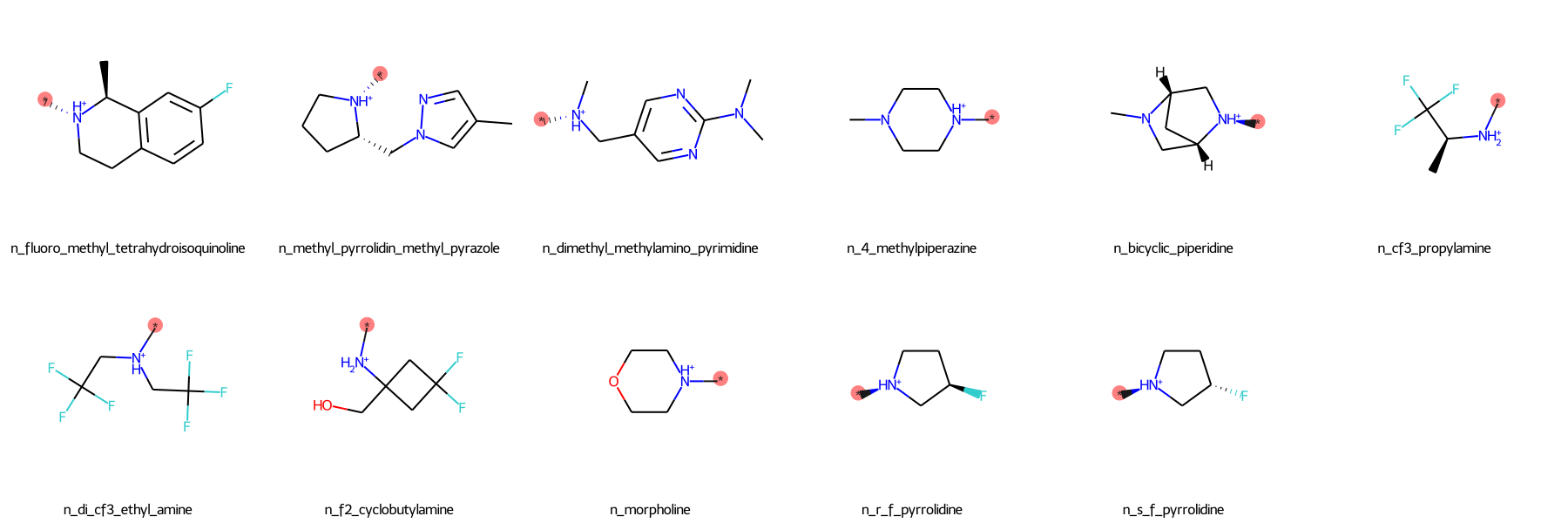
The SilcsBio GUI provides a wide array of ligand modifications. The readily available ligand substitution modifications are listed below. In the images below, the red circle and asterik indicate the location at which the substitution will be added to the input ligand. In addition to these substitutions, please see the “Repl.” tab within the GUI itself for readily available replacement options.
Small Substituents¶
Moderate Substituents¶
Ligand Modifications Using the CLI¶
To perform chemical group transformations using the CLI, you will need to create
a text file modifications.txt with instructions
listing the desired modifications to your parent ligand.
Three types of modifications can be performed, as listed in the table.
Command |
Modification |
|---|---|
JOIN |
Join a fragment (mol2) to the parent at a defined site |
REPL |
Replace an atom at a defined site on the parent with a fragment (mol2) |
MUDE |
Change an atom at a defined site on the parent to another atom-type |
An example ligand modification input file is povided with your SilcsBio
server software, ${SILCSBIODIR}/examples/ssfep/modification.txt.
For details on how to modify and process this file to create Mol2 files containing modifications to your parent ligand, run the following command:
${SILCSBIODIR}/programs/chemmod -h
Your SilcsBio server software comes with a large number of fragments in Mol2 format that can be added/joined to your parent ligand. These fragments are found in:
${SILCSBIODIR}/data/groups
A JOIN operation can be performed between the parent and a ch4.mol2
fragment by adding the following line to modifications.txt:
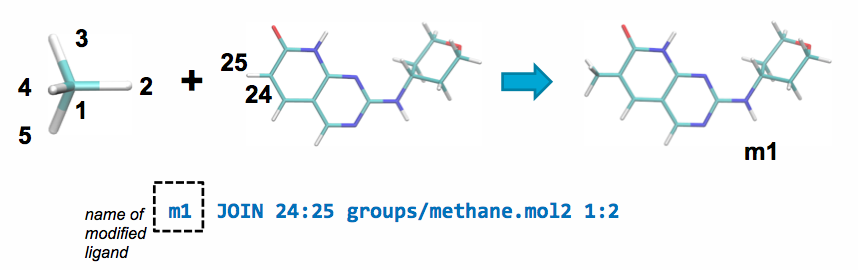
This line will join atom 24 in the parent ligand with
atom 1 in methane.mol2 and delete atoms 25 and 2 in the parent ligand and
the joined fragment, respectively.
A REPL operation can be performed between the parent and an nh2.mol2
fragment by adding the following line to modifications.txt:
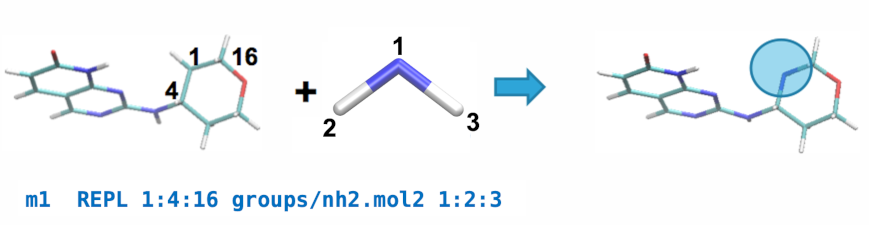
This line will replace atom 1 of the parent ligand with atom 1 of the fragment by aligning atoms 2 & 3 of the fragment with atoms 4 & 16 of the parent, respectively, and replacing the carbon in the ring with a nitrogen atom.
The same transformation in the previous section can also be achieved using a MUDE operation:
m2 MUDE MU 1:7 DE 21
This line will mutate atom 1 (atom index number) in the parent with nitrogen (atomic number 7) along with deleting the hydrogen (atom index number 21) attached to the parent carbon.
To evaluate multiple modifications to a single site on the parent ligand, use the following syntax:
m1 JOIN 24:25 <filename>
Replace <filename> with the name of a text file containing each of the
multiple modifications, with one modifications per line. Examples of such
text files are list.txt and small_join_list.txt located in the
${SILCSBIODIR}/data/ folder, and you can use these example files as
the basis for your own custom file that includes modifications using Mol2
files from ${SILCSBIODIR}/data/groups or your own custom Mol2 files.
Be careful to pay attention to chemistry; if a modification in your text
file is not suitable for a site, you can comment it out using ! at
the beginning of that line. Based on the above JOIN line, Mol2 output
files from running ${SILCSBIODIR}/programs/chemmod will all begin
with the prefix m1_.